Abstract
Alkanes are major constituents of plant-derived waxy materials. In this study, we investigated the abundance, community structure and activity of bacteria harbouring the alkane monooxygenase gene alkB, which catalyses a major step in the pathway of aerobic alkane degradation in the litter layer, the litter–soil interface and in bulk soil at three time points during the degradation of maize and pea plant litter (2, 8 and 30 weeks) to improve our understanding about drivers for microbial performance in different soil compartments. Soil cores of different soil textures (sandy and silty) were taken from an agricultural field and incubated at constant laboratory conditions. The abundance of alkB genes and transcripts (by qPCR) as well as the community structure (by terminal restriction fragment polymorphism fingerprinting) were measured in combination with the concentrations and composition of alkanes. The results obtained indicate a clear response pattern of all investigated biotic and abiotic parameters depending on the applied litter material, the type of soil used, the time point of sampling and the soil compartment studied. As expected the distribution of alkanes of different chain length formed a steep gradient from the litter layer to the bulk soil. Mainly in the two upper soil compartments community structure and abundance patterns of alkB were driven by the applied litter type and its degradation. Surprisingly, the differences between the compartments in one soil were more pronounced than the differences between similar compartments in the two soils studied. This indicates the necessity for analysing processes in different soil compartments to improve our mechanistic understanding of the dynamics of distinct functional groups of microbes.
Keywords: alkane monooxygenase, microbial community function, plant litter, soil compartments, waxes
Introduction
Soil harbours a multitude of different microhabitats for (micro)-organisms that form the basis for the vast diversity and functional heterogeneity often found in soils (Curtis and Sloan, 2005). Such habitats are characterised by multidimensional interfaces at the μm to mm scale, and are the result of the interplay of soil microbiota with their physical and chemical environment (Totsche et al., 2010). As these structural elements are highly dynamic in response to the surrounding environmental conditions, they can be considered as hotspots for microbial activity and the basis of all ecosystem services provided by soils (Yaalon, 2000). In particular, the degradation of polymeric substances and other complex molecules might be depending on the structure of microhabitats either due to present varying microbial communities, different sorption properties and/or oxygen and nutrient availability. Such heterogeneity remains undetected, if vast amounts of bulk soils are sampled with a soil auger, homogenised and divided into subsamples for analysis.
Alkanes are major compounds of plant leaves (Eglinton et al., 1962). The corresponding cuticulary waxes are produced as protective systems against phytopathogens or to minimise water loss by uncontrolled evaporation (Avato et al., 1990; Gniwotta et al., 2005). Compound concentration and composition of these waxes vary between plants, thus allowing differentiation of several plant families or plants with different metabolic properties like C3 and C4 plants (Rieley et al., 1993; Collister et al., 1994; Maffei, 1996). Earlier studies revealed the release of plant wax-derived alkanes during litter degradation and their transport into the surrounding soil (Lichtfouse et al., 1994; Cayet and Lichtfouse, 2001; Dignac and Rumpel, 2006). Mainly alkanes with a chain length shorter than C20 can be transported into deeper soil layers and enter different soil compartments, where their bioavailability can be drastically reduced by physico-chemical mechanisms such as (i) adsorption to soil organic matter and clay minerals (Manilal and Alexander, 1991; Richnow et al., 1995), (ii) adsorption to non-aqueous-phase liquids and (iii) diffusion into smaller pores inaccessible for microbes (Bosma et al., 1996). Soil microbes may also influence the bioavailability of alkanes. For example, bacteria can actively promote bioavailability by reducing their distance to these substrates by chemotaxis (Lanfranconi et al., 2003), changing their cell wall properties (de Carvalho et al., 2009) or producing biosurfactants such as rhamnolipids or the bioemulsifying protein. These processes facilitate bacterial attachment to substrate reservoirs (Wick et al., 2000) and transfer of hydrocarbons into aqueous phases (Holden et al., 2002). Beside alkane bioavailability other biotic and abiotic parameters like the presence of dissolved organic carbon (Smith et al., 2009), the access to nutrients or the amount of oxygen present influence alkane biodegradation in different soil compartments.
The capability of alkane catabolism via aerobic and anaerobic degradation pathways is found among a broad range of fungi and bacteria. Although the chemically inert alkanes are activated by the addition of fumarate (Heider et al., 1999; Widdel and Rabus, 2001; Wilkes et al., 2003) during anoxia, aerobic activation is accomplished by the terminal (Sepic et al., 1995; Koma et al., 2001; Van Hamme et al., 2003) or subterminal (Whyte et al., 1998; Kotani et al., 2006, 2007) introduction of oxygen. For alkanes with a chain length <C30, which can be considered as typical for plant waxes, terminal oxygen introduction is mainly catalysed by the membrane bound, rubredoxin-dependent di-iron alkane monooxygenase (AlkB), which is found among Actinobacteria, α-, β- and γ-Proteobacteria (van Beilen et al., 2003; van Beilen and Funhoff, 2007).
In this study, we report on the dynamics of the abundance, activity and community structure of microbes harbouring the alkane monooxygenase gene alkB during the degradation of litter-derived alkanes in different soil compartments (litter layer; litter–soil interface; bulk soil). We postulate that in soil compartments close to the litter material, the type of litter material highly influences abundance, activity and community structure of alkB harbouring bacteria. With increasing distance from the litter layer, the influence of the soil type should increase. For this, intact soil cores of two arable soils, differing significantly in their composition and texture, were incubated with litter of maize and pea. These two plants were chosen as they represent different assimilation pathways (C3 and C4 plants, respectively) and contain different alkane concentrations as well as different compositions (Avato et al., 1990; Gniwotta et al., 2005). The different compartments were analysed after 2, 8 and 30 weeks of incubation. The alkB gene copy numbers and amounts of alkB transcripts were quantified using SybrGreen-based real-time PCR, whereas the community structure of alkB harbouring microbes and the corresponding transcripts was determined using terminal restriction fragment polymorphism (T-RFLP) analysis. Furthermore, the amount and composition of litter-derived alkanes were determined in the three different compartments.
Materials and methods
Soil sampling and microcosm experiment
A total of 96 soil cores (diameter 5 cm; height 5 cm) were taken with a soil auger from two different top soils (0–5 cm; 48 cores each) at an agricultural research farm located 45 km north of Munich (www.helmholtz-muenchen.de/scheyern) in autumn 2008 after the harvest of winter wheat. The soil texture of the silty (60.6% silt, 18% sand, 21.4% clay) and the sandy soil (31.4% silt, 55.2% sand, 13.4% clay) was determined in 2003 after Sinowski and Auerswald (1999). Both soils had a pH of 6.0 after extraction with 0.01 M CaCl2. The soil cores were equilibrated for two weeks at 14 °C and 55% of the maximum water holding capacity to reset soil microbial activity. Three cores of each soil type were sampled after equilibration and served as time point zero treatments (T0). Nine soil cores of each soil type were incubated with 1 g dry weight (dw) fresh litter material from maize (Zea mays L), respectively, pea (Pisum sativum L) with a piece size of 25 mm2, which was applied carefully on the top of the soil core. Additionally, for each litter and soil type nine cores were incubated using litter material in litterbags (1 g litter material per bag; 50 μm mesh size of the bags), which were carefully covered by soil to determine litter degradation rates. Nine cores of each soil type without litter addition served as controls. All cores were incubated in the dark up to 30 weeks at a constant temperature of 14 °C and soil moisture of 55% of maximum water holding capacity.
Sampling of three soil cores per treatment (sand-pea; sand-maize; sand-control; silt-pea; silt-maize; silt-control) was performed 2 (T1), 8 (T2) and 30 (T3) weeks after litter addition. For each sampling three compartments (litter layer, litter–soil interface and bulk soil 1–1.5 cm below the interface) were sampled from each core and treated as true replicates from the treatments. To obtain the litter–soil interface, the remaining litter material was carefully removed with a pair of forceps and then carefully sampled using a clean spatula up to a depth of 2–3 mm below the litter layer. Samples were either shock frozen in liquid nitrogen and stored at −80 °C for molecular analysis or, respectively, dried with anhydrous Na2SO4 (ca 10–15 g) for the analysis of alkane amount and composition. For the determination of litter degradation rates, litter bags were removed from the soils of the corresponding treatments at T1, T2 and T3, and the air-dried litter material quantified gravimetrically.
Alkane analysis
Abundance and composition of alkanes were determined in triplicate from either 1.5 g soil or 0.5 g of pooled litter. Samples were extracted twice with 20 ml (60 min extraction time) and 10 ml (30 min) hexane (HPLC-grade; Merck, Darmstadt, Germany) using a horizontal shaker at room temperature, and then concentrated to 1 ml using a Turbo Vap II vacuum system (Zymark, Idenstein, Germany). In order to correct potential alkane losses during extraction, 50 ng of deuterated hexadecane per ml of hexane (Dr Ehrenstorfer GmbH, Augsburg, Germany) was used as extraction standard. Litter extracts were split into two fractions of 0.5 ml, where each of the fractions was further purified by using silica gel columns containing 0.5 g of cleaned and activated silica 60 (35–50 mesh, Merck) and 3–5 ml of hexane (HPLC-grade; Merck). Joined extracts of the columns were concentrated (Turbo Vap II vacuum system) to 0.5 ml. Gas chromatographic mass spectrometry (GC-MS) measurements of concentrated extracts were performed with an HP 6890 gas chromatograph (Hewlett Packard, Palo Alto, CA, USA), which was equipped with an HP-5 MS column and an HP 5973 mass spectrometer for detection (both Agilent Technologies, Santa Clara, CA, USA). Final temperature was set to 320 °C, at a final time of 7 min and an initial helium flow of 0.8 ml min−1. For the MSD Transfer Line Heater an initial temperature of 280 °C was used.
Extraction of nucleic acids and cDNA synthesis
DNA and RNA were co-extracted as recently described (Töwe et al., 2011) and subsequently separated using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Quality and quantity of isolated nucleic acids were determined spectrophotometrically at 260 and 230 nm (Nanodrop ND-1000, Peqlab, Erlangen, Germany). First strand cDNA was synthesised with SuperScript II Reverse Transcriptase (Invitrogen, Darmstadt, Germany) using 1 pmol of random hexamer primer.
Quantification of alkB genes and transcripts
SybrGreen-based quantitative real-time PCR (qPCR) was performed with the ABI Prism 3300 (Applied Biosystems, Foster City, CA, USA) system for the DNA and cDNA. PCR reaction mixtures with 25 μl reaction volume contained 2 mM magnesium chloride, 0.1 μM of alkB specific primers (Table 1; Kloos et al., 2006), 0.06% BSA, 1 × Power Sybr Green Master Mix (Applied Biosystems) and 5–15 ng DNA or 10–40 ng cDNA, respectively. The qPCR was carried out as previously described (Pérez-de-Mora et al., 2010) using a cloned fragment of alkB (550 bp) from Pseudomonas putida as standard.
Table 1. Primers used for the quantitative PCR and T-RFLP analysis.
| Primer | Sequencea,b | Reference |
|---|---|---|
| alkB-1f | 5′-AAYACNGCNCAYGARCTNGGNCAYAA-3′ | Kloos et al. (2006) |
| alkB-1r | 5′-GCRTGRTGRTCNGARTGNCGYTG-3′ | Kloos et al. (2006) |
Underlined positions were substituted by an inosine nucleotide to improve amplification efficiency.
N bases A, T, C or G; Y bases C or T; R bases G or A.
Standard curves were linear (r2>0.99) over five orders of magnitude and amplification efficiency was around 86%. No fluorescence signals were detected in the non-template controls. Signal acquisition at 78 °C impeded biased fluorescence signal detection due to primer dimers.
Community structure of alkB harbouring bacteria and their transcripts
DNA and cDNA were used for T-RFLP analysis to track changes in the diversity of alkB harbouring bacteria on DNA as well as mRNA level. Briefly, PCR reaction mixtures with 25 μl volume were composed of alkB specific primers (Table 1), with alkB_1f labelled with 6-carboxyfluorescein (6-FAM), 15–20 ng DNA or 120 ng cDNA, 1 × Taq PCR Master Mix Kit (Qiagen) and 0.12% BSA. The annealing temperatures were set to 58.7 °C for 30 cycles for DNA and 58 °C for 35 cycles in the case of cDNA. PCR reactions were accomplished with no further changes in the PCR protocol described in the previous section. Afterwards, amplification products were purified with Wizard SV Gel and PCR Clean-UP System (Promega, Mannheim, Germany) according to the manufacturer's instructions and subsequently quantified spectrophotometrically at 260 nm (Nanodrop ND-1000, Peqlab). An amount of 100 ng purified DNA-derived PCR product or 10–50 ng of cDNA-derived PCR product, respectively (pooled from three independent PCR reactions), was digested at 37 °C over night using two units of HpyCH4V (New England BioLabs, Beverly, MA, USA). After precipitation of the restriction fragments with ethanol, T-RFLP was run in an ABI PRISM 3100 genetic analyzer system (capillary injection time of 15 s) using GeneScan 500 ROX as size standard (both Applied Biosystems). Data were analysed with GeneMapper V3.7 software (Applied Biosystems).
Statistical analysis
Multifactorial ANOVA calculations were run with log-transformed data. For statistical analyses of the abundance of the alkB genes and transcripts as well as the community structure of alkB harbouring bacteria, the statistical software R version 2.10.0 (R Development Core Team, 2009) was used. Normal data distribution was checked by using histograms and the Kolmogorov–Smirnov test at a level of significance of P⩽0.05. See details on the statistical analysis of T-RFLP data in the Supplementary Material S1.
qPCR data were calculated on the basis of the amount of soil or litter material. The alternative to relate functional gene abundance to 16S rRNA gene abundance has been not feasible, as a shift from r- to k-strategists may occur during the incubation time that excludes the use of constant conversion factors, because r- and k-strategists are known to contain different rRNA operon copy numbers (Klappenbach et al., 2000).
Results
Litter degradation rates
Litter degradation rates based on the litter bag experiments performed followed a logarithmic function as described previously (Wang et al., 2004; Ha et al., 2007). The decay of pea litter was significantly faster than the degradation of maize litter (P<0.001) independent of the soil type. Whereas after 2 weeks of incubation only half of the initial maize litter amount remained, nearly 80% of the initial pea litter was degraded. The soil type did not significantly influence the litter decay (Supplementary Materials S2 and S5).
Alkane analysis in the different compartments
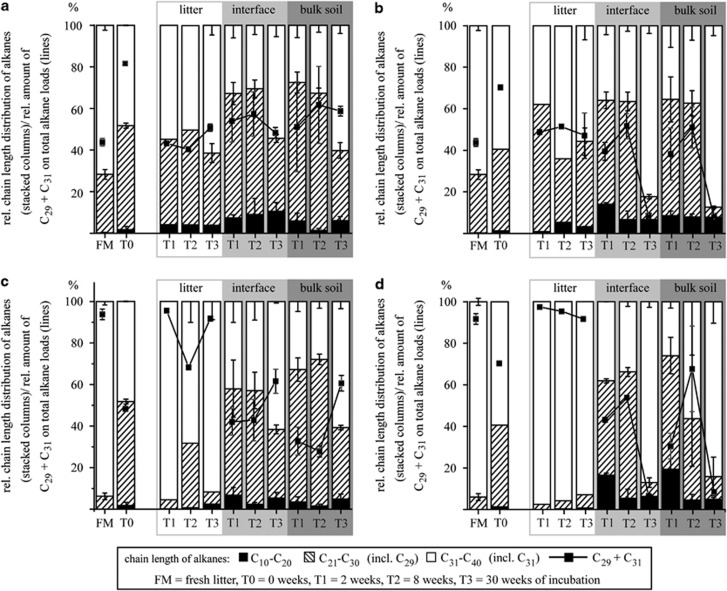
Even- and odd-numbered, straight-chain alkanes of chain-lengths between C10–C40 as well as pristane and phytane were quantified in all compartments (litter layer; litter–soil interface and bulk soil) at all sampling time points (Table 2 and Figure 1). Alkane amounts in plant litter exceeded the alkane loads of the underlying soil compartments by two to four orders of magnitude. Fresh plant material comprised alkane contents of about 180 μg g−1 for maize litter and 6500 μg g−1 for pea litter (Table 2). Alkane degradation rates were comparable in all litter samples with 43–87% of the total alkane concentration being lost within the first 8 weeks of incubation. Independent of the soil type (P>0.1), maize and pea litter differed significantly (P<0.05) in their relative chain-length distribution over time with considerably higher fractions of very long chained alkanes (C31–C40) in pea litter. Nonacosane (C29) and hentriacontane (C31), which are known to prevail in maize and pea (Avato et al., 1990; Gniwotta et al., 2005), constituted between 76–97% of the total alkane content in pea litter and 40–62% of the total alkanes in maize (Figure 1).
Table 2. Total alkane concentration at the three analysed compartments during the incubation of sandy and silty soil microcosms with maize and pea litter.
| Compartment | Time |
Maize |
Pea |
||
|---|---|---|---|---|---|
| Sandy soil | Silty soil | Sandy soil | Silty soil | ||
| Litter (μg g−1) | FM | 183.4 (22.0) | 183.4 (22.0) | 6544.8 (2013.4) | 6544.8 (2013.4) |
| T1 | 29.9 (ND) | 169.3 (ND) | 7524.3 (ND) | 6021.0 (ND) | |
| T2 | 26.3 (ND) | 36.4 (ND) | 881.3 (ND) | 3705.9 (ND) | |
| T3 | 25.4 (ND) | 56.6 (ND) | 2111.4 (ND) | 1428.5 (ND) | |
| Interface (0-1 mm) (ng g−1) | T1 | 52.0 (13.0) | 46.0 (2.0) | 52.0 (16.0) | 50.0 (6.0) |
| T2 | 39.0 (15.0) | 36.0 (1.0) | 66.0 (31.0) | 47.0 (4.0) | |
| T3 | 49.0 (12.0) | 105.0 (29.0) | 81.0 (17.0) | 96.0 (27.0) | |
| Bulk soil (ng g−1) | T0 | 167.0 (52.0) | 206.0 (ND) | 167.0 (52.0) | 206.0 (ND) |
| T1 | 67.0 (26.0) | 52.0 (6.0) | 78.0 (7.0) | 40.0 (5.0) | |
| T2 | 128.0 (6.0) | 39.0 (9.0) | 71.0 (3.0) | 48.0 (14.0) | |
| T3 | 120.0 (40.0) | 74.0 (2.0) | 79.0 (22.0) | 108.0 (24.0) | |
Sampling time points for bulk soil were at the beginning (T0) and after 2 (T1), 8 (T2) and 30 (T3) weeks of incubation. Besides the fresh litter material (FM), litter has been also analysed at T1–T3. Samples from the soil–litter interface have been analysed from time points T1–T3. Note that concentrations are given in μg g−1 (litter samples) or ng g−1 (soil samples), respectively. Standard deviations are given in brackets (n=3). As the amount of remaining litter material was too low for single measurements at T1, T2 and T3 litter material was pooled from the three replicates; thus no standard deviation is given (ND).
Figure 1.
Relative distribution of medium- (C10–C20), long- (C22–C30) and very long-chain (C32–C40) alkanes and for the dynamics of nonacosane and hentriacontane (C29 and C31) as control for the most abundant plant wax alkanes during the incubation of sandy (a, c) and silty (b, d) soil microcosms covered with litter of maize (a, b) or pea (c, d). The litter layer, litter–soil interface (0–1 mm) and the bulk soil are indicated. Error bars represent standard deviations (n=3).
Similar concentration dynamics were monitored for the different soil compartments in both soil types with no significant influence (P>0.1) of the litter type over time (Table 2). Starting with comparable alkane loads in silty and sandy soil (206 ng g−1 soil, respectively, 167 ng g−1), the total alkane concentrations decreased to 20–47% of the initial concentration after 2 weeks (T1) in all soil samples. The differences in the residual amounts at T1 were significantly dependent on the soil type (P<0.05) but independent of the soil compartment (P>0.1). Until the end of the experiment, further significant changes in total alkane amounts were observed only for bulk sandy soil incubated with maize at T2 (8 weeks) as well as both litter–soil interface and bulk soil compartments from the silty soil treatments at T3 (30 weeks) where up to three times higher alkane loads (P<0.05) were monitored compared with the preceding time point.
Changes in the relative chain-length distribution of alkanes in soil were significantly influenced by the soil type and additionally by the incubation time (P<0.05). Although sandy and silty soils shared a similar distribution of alkanes with different chain-lengths at T0 (2% 50% 48%, respectively, 1% 40% 59% for C10–C20; C21–C30; C31–C40) with nonacosane and hentriacontane constituting 82 and 70% of the total alkane content, pronounced differences were observed at T3. Whereas moderate changes of chain-length distribution patterns were observed during the incubation in sandy soils only, long-chained alkanes (C31–C40) represented the dominating alkane fraction (85%) in silty soils at T3. At this time a highly significant impact of the soil type was monitored (P<0.001) while the litter type had no influence. Interestingly, only the relative amount of nonacosane and hentriacontane was influenced by the litter type at this time (P<0.05), which is coincident to earlier results (Avato et al., 1990; Gniwotta et al., 2005).
Quantification of alkB genes and transcripts in the different compartments
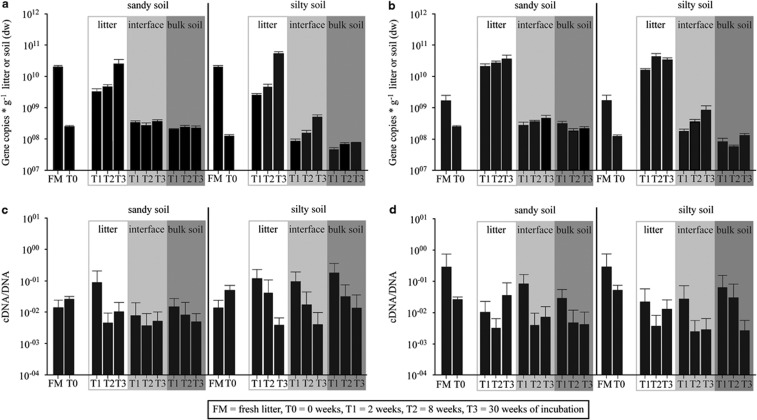
All results of the quantification of alkB genes and transcripts are summarised in Figure 2. The initial alkB gene copy number differed in both litter types significantly (P<0.05). While in maize litter 3 × 1010 copies g−1 litter (dw) were monitored in the fresh litter material, gene copy numbers in pea litter were one order of magnitude lower. In the litter layer significant interactions between incubation time and litter type (P=0.001) reflect the influence of both factors on alkB abundance. Although alkB gene copy numbers dropped at T1 and increased again towards T3 in all maize litter samples, in pea litter a significant increase in alkB copy numbers was mainly visible at the first 2 weeks of incubation towards T1. After 30 weeks of incubation alkB gene copy numbers were comparable in both litter types and reached almost 1 × 1011 copies g−1 litter (dw). The soil type had no significant influence on alkB gene copy dynamics in litter. AlkB gene copy numbers at the litter–soil interfaces and in the bulk soil compartments were significantly lower (P<0.001) than in the respective litter layer and ranged between 2 × 108 and 9 × 108 copies g−1 soil (dw). In silty soil samples a clear reduction of alkB copy numbers from the litter–soil interface towards the bulk soil was visible. Over the time, gene copy numbers at the litter–soil interface increased, whereas no significant changes could be detected in bulk soil independent of the type of litter. In contrast, in the sandy soil samples no differences were monitored between both soil compartments and alkB gene copy numbers remained stable in the litter–soil interface over time.
Figure 2.
Changes in the abundance patterns (a, b) and the specific induction of the alkB gene (c, d) in the litter layer, the litter–soil interface and bulk soil during the incubation with maize (a, c) and pea (b, d) litter. The specific induction is referred to the ratio of alkB mRNA and DNA. Error bars represent standard deviations (n=3).
Overall copy numbers of alkB transcripts followed the same trend like described above (data not shown). To describe specific transcription rates of alkB in the different compartments the ratio between mRNA and DNA copy numbers were calculated. Specific induction of alkB transcripts could be clearly observed independent from the soil type (P=0.95) at the maize litter compartment within the first 2 weeks of incubation followed by a remarkable decrease. In contrast, highest specific induction rates for alkB-related transcripts were found in the fresh pea litter and decreased mainly in the first 2 weeks of incubation towards T1. In the silty soil samples treated with maize litter a clear influence of the litter addition was visible on the specific induction rates of alkB in the litter-soil compartment as well as in the bulk soil compartment after 2 weeks of incubation. In all other treatments, the addition of the litter material did not increase specific transcription rates in the two soil compartments towards T1. At T2 and T3, specific induction rates of alkB genes were reduced in all treatments compared with T0 independent of the plant litter type, the soil compartment analysed or the soil type (P=0.873).
Gene and transcript copy numbers of alkB did not change significantly over time in control microcosms without litter addition (data not shown).
Community structure of alkB harbouring bacteria and their transcripts in the different compartments
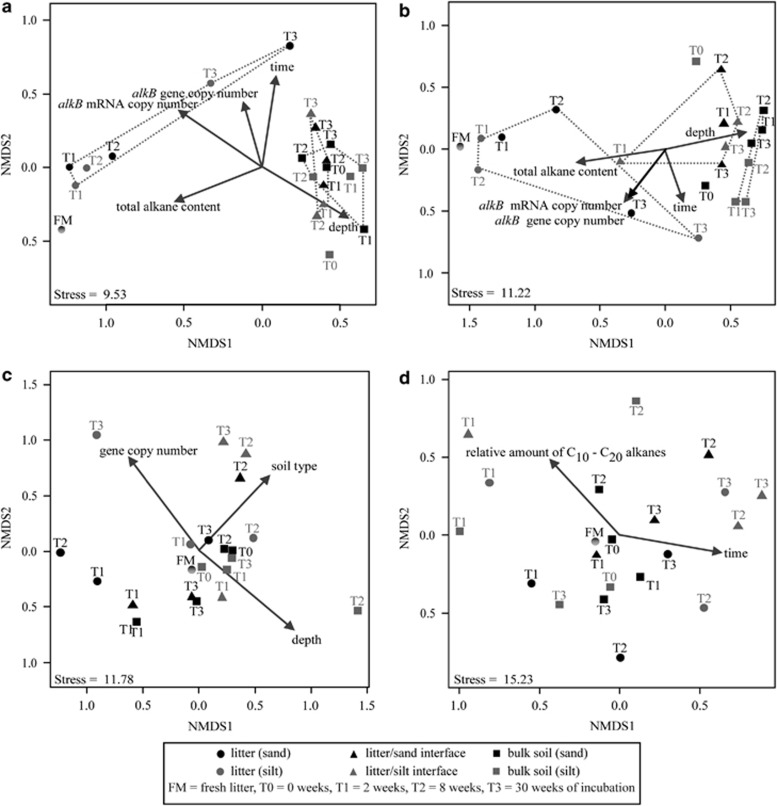
Two weeks after the litter addition, an increase of total T-RF numbers (that is, richness of alkB genes) was observed in the litter-soil and bulk soil compartment irrespective of the soil type (Supplementary Material S7). Nonmetric multidimensional scaling plots calculated on the basis of Bray–Curtis similarity indices (including presence and relative abundance of T-RF) displayed a strong differentiation between communities on the DNA level in response to the sampled compartment, the litter type and the incubation time (Figure 3; Supplementary Material S4). Overall, alkB harbouring communities in the litter–soil interface as well as in the bulk soil were more related to each other than to communities colonising the litter material. For example, T-RF 63, 117 and 475 were detected in most DNA samples from soil, but were far less frequent in litter samples. Conversely, the T-RF 260, 324 and 401 were far more specific for litter samples and occurred in only few soil samples (Supplementary Material S8) if at all. Interestingly, although less frequent in the litter samples based on DNA analysis, T-RF 63 and 117 were dominant members of the T-RF profiles of the mRNA samples, indicating high transcription rates of the corresponding genes at least at the time point of sampling. The analysis of samples derived from replicate treatments did not reveal significant differences in T-RFLP pattern (data not shown); therefore data in Figure 3 and others is based on analysis of pooled samples from replicated treatments for clarity reasons.
Figure 3.
T-RFLP based NMDS plots of alkB gene harbouring communities based on DNA-derived T-RFLP analysis (a, b) and mRNA-derived T-RFLP-analysis (c, d) in sandy and silty soil microcosms covered with maize (a, c) and pea litter (b, d). Community similarity was calculated using Bray–Curtis similarity measurement, which includes presence and relative abundance of T-RF. Note that only bulk soil was sampled at T0, as no differences between litter–soil interface and bulk soil were expected at the beginning of the incubation. Arrows are correlation vectors of diversity differences and environmental conditions with significance factors P<0.1 (for exact significance level see Supplementary Material S4). Monte–Carlo permutation model based analysis with 1000 permutation and final ANOVA was applied to test for significance.
Nonmetric multidimensional scaling plots of amplified mRNA displayed no differentiation among communities in the different compartments, as observed on DNA level, irrespective of the litter material (Figures 3c and d). However, the composition of transcripts of alkB based on T-RF analysis was significantly impacted by the soil type in soil cores incubated with maize litter, whereas incubation time and the relative amount of C10–C20 influenced the alkB genes and alkB transcripts significantly in cores incubated with pea litter (Supplementary Material S4).
Based on ongoing cultivation studies using samples from the same experiment, several of the T-RF could be tentatively assigned to T-RF obtained from alkB sequences originating from soil and litter isolates (Supplementary Material S3). For example, T-RF 324 and 459 were specific for Actinomycetales-related isolates harbouring alkB sequences homologous to the alkB sequence types of Rhodococcus sp. Q15 alkB2 (AF388182) and Gordonia sp. TF6 (AB112870), respectively. These T-RF were more common in DNA from litter samples and were, if at all, only detectable in soil samples incubated with litter, but never in controls.
A total analysis with nonmetric multidimensional scaling of T-RFLP patterns of all treatments and layers (including controls) on genomic and transcriptomic scale revealed differentiation between the alkB harbouring and alkB expressing communities (Supplementary Material S9). The alkB harbouring communities especially from the litter–soil interface and bulk soil (irrespective of the soil type) shared more similarity to each other (domination sector III) than to the respective alkB expressing communities (mainly sectors II) (molecule type, P<0.001). This indicates a high similarity to litter communities expressing alkB (sector II). The litter type had significant influence on the differentiation between alkB harbouring and expressing communities of all samples including controls (P<0.001) as also observed for the soil type (P<0.05) and the vicinity to the litter (depth, P<0.001). Control samples especially of bulk soil incubated without litter had similar patterns for alkB genes and transcripts (mainly sector I) and differed from soil samples incubated with litter (dominating sector III). Overall, the community structure of alkB harbouring bacteria genes and their transcripts in the non-litter controls did not change (Supplementary Materials S6 and S9).
Discussion
In this study, we related gene copy numbers, structure and activity of a functional community in different soil compartments to the presence of the corresponding substrate. We could demonstrate that the investigated bacterial populations are highly dynamic in the different soil compartments, which have been investigated and at least partly controlled by substrate availability.
Surprisingly litter application resulted only in a growth of alkB harbouring bacteria in the litter layer of soils where pea litter has been added. This increase in biomass could not be related to the degradation of alkanes only, as the amounts in litter are too low. Therefore, the use of other plant-derived compounds by the alkane degraders and co-metabolic degradation must be postulated. This hypothesis is also verified by the fact that in pea litter, which is easily degradable compared with maize litter and nutrient release is much faster, a steady increase of alkB harbouring bacteria could be observed. In the litter–soil interface in silty soils also an increase in alkB harbouring bacteria could be detected at T3 independent from the litter type. This observation might be related to the fact that leaching of alkanes is much slower in silty soils compared with sandy soils as indicated by the higher alkane concentrations in silty soil samples at T3; this prolonged substrate availability in silty soil samples obviously stimulated the growth of alkB harbouring bacteria.
Generally, prolonged incubation time of the soil cores with the litter material resulted in distinct changes in alkB expression and diversity of alkB harbouring bacteria. These shifts as well as the increased abundance were most pronounced in the litter layer likely indicating that significant parts of the available alkanes were readily degraded. This fact is supported by data from earlier studies, where extensive community structure changes of alkB harbouring bacteria in nutrient-enriched soils as compared with control plots over time has been described in several field studies (Röling et al. 2004; Hamamura et al. 2006; Vázquez et al., 2009).
Substrate loads and relative chain-length distribution of alkanes in the compartments were significantly responsible for the distinct T-RFLP alkB fingerprints from litter and soil on the DNA level. These strong differences reflect changes in the presence as well as in the relative abundance of T-RF in litter and soil. This might indicate that alkane degraders in the litter layer were specific to or possibly introduced with the plant material rather than being part of the soil microbial community.
However, on the expression level the similarity in the number of transcripts between litter and soil samples indicates that external environmental conditions rather than the high abundance of certain degraders are responsible for shaping the community structure of active alkane degraders harbouring the alkB gene. Furthermore, specific alkB gene paralogues could be expressed similarly in litter and soil in response to the comparable growth conditions. The paralogues functions are yet unclear, but it was hypothesised that they are preferentially expressed depending on the chain-length of the available alkanes or on the growth state of bacteria (Marin et al., 2001; van Beilen et al., 2003; Amouric et al., 2010).
Alkanes released upon decay of litter material are possibly solubilised by surface-active agents (Holden et al., 2002), facilitating their transport into the surrounding soil, for example, with flowing water (Lichtfouse et al., 1994). Interestingly, total alkane loads in the litter–soil interface decreased significantly rather than increased, after litter amendment. Together with the stable numbers of alkB genes and transcripts in some soil compartments, this could support degradation activity of alkane degraders relying on enzyme systems others than AlkB (for example, Cyp153). In contrast, elevated alkB gene and transcript numbers in bulk soil compartments were detected after litter amendment, thus hinting at a concomitantly stimulated alkane degradation activity and a fast consumption of the substrate by alkB harbouring bacteria.
Soil type dependent differences in the bioavailability of alkanes might occur due to the different porosities and mineral compositions of sandy and silty soils. In fact, the macroporosity of sandy soils, as opposed to the microporosity and high clay content of silty soils, may favour the transport of alkanes (Galin et al., 1990; Arthurs et al., 1995) and therefore influence the structure and activity of alkB harbouring bacteria differently. At T3, the strong soil type dependent differences in the relative chain-length distribution and a concomitantly higher total alkane amount in silty soil were accompanied by elevated alkB gene numbers in the interface compartments of this soil type. Soil texture induced alkane sequestration, hence could have been responsible for a reduced ad hoc bioavailability and concomitantly retarded substrate consumption in silty soil.
The present study, interestingly, could not verify any influence of the soil type on the community structure of bacteria harbouring the alkB gene, as they were remarkably similar between sandy and silty soils, whereas simultaneously expressing similar alkB gene abundance and expression. In contrast, the differences in the community structure of alkB harbouring bacteria were much higher in the soil compartments within one soil type than in the same soil compartments between the two soil types. This points to a distinct influence of the plant litter on the community structure of potential degraders that is more pronounced in communities located closer to the litter layer. Factors such as the availability of nutrients or electron acceptors introduced by the plant litter may accordingly be important (Wiesenberg et al., 2004; Dignac and Rumpel, 2006) forming steep gradients and thus preferentially influencing the community structure. Furthermore, plant litter-derived degraders could be transferred to the litter–soil interface along fungal hyphae (Kohlmeier et al., 2005), without reaching the deeper soil layers.
However, on the expression level, the depth differentiation between the two soil compartments was less pronounced as also observed for the alkB transcripts in litter and soil. This and the dependency of the community structure of bacteria expressing the alkB gene in pea litter incubated samples on the relative amount of medium-chain alkanes (C10–C20) further supports the hypothesis of preferentially expressed alkB gene paralogues. The changes in the alkB expression patterns of maize litter incubated samples significantly correlated with the soil type, which had not been observed on the DNA-level and could thus be explained by the soil type-dependent relative chain-length distribution of the alkanes. At T3, the higher fraction of very long-chain alkanes in silty soil (C31–C40) at both compartments possibly induces alkB gene paralogues others than expressed with long-chain alkanes (C21–C30), which were more abundant although not dominant in sandy soil. Furthermore, the significant differences in the relative amounts of nonacosane and hentriacontane between the soil types might have a role.
Conclusion and outlook
The observed dynamics of abundance, community structure and activity of functional communities are often overlooked when soil samples are taken with a soil auger and further processed by mixing and sieving. However, to understand the functionality of soils in relation to the performance of microbial communities, the spatial resolution of analyses close to biogeochemical interfaces as they have been defined by Totsche et al. (2010) have to be improved. These authors stated that ‘linking the heterogeneous architecture of soil interfaces with the diverse structure of microbial communities in predictive ways in order to understand soil functions and the role for ecosystem services has yet to be seen as one of the grand decadal challenges in science'. The structural and compositional heterogeneity of these interfaces is one likely reason for the vast diversification of microbes (Curtis and Sloan, 2005; Gans et al., 2005). Biogeochemical interfaces may stimulate the metabolic activity and are supposedly sites for horizontal gene transfer (Top and Springael, 2003; van der Meer and Sentchilo, 2003). This role of interfaces has also been demonstrated impressively using model biofilms (Molin and Toker-Nielsen, 2003). Therefore, future studies must address even smaller scales than those addressed by the present study to get more mechanistic insight into the functioning of biogeochemical interfaces and their dynamics. There is a particular need to employ minimally destructive sampling techniques and in situ analysis to obtain an unbiased view of the ecology of communities at high resolution.
Obviously in our study litter type-specific abundance of alkB and its expression patterns thereby seem to reflect not solely the different bioavailability of alkanes but might be also the result of the succession of alkB harbouring bacteria by alkane degrading fungi (for example, Fusarium sp, Aspergillus sp, Penicillium sp). These fungi are known as important drivers of litter degrading communities and likely competitors with the bacteria for the same substrate. These alkane-degrading microbes do not use the alkane monooxygenase pathway for alkane degradation and therefore cannot be tackled by using alkB specific primers. Therefore, to describe total functional communities involved in the degradation of plant-derived alkanes abundance, the community structure and the activity pattern of those microbes need to be described in future experiments, for example, by using metagenomic analysis and expression cloning approaches.
Finally T-RFLP analysis is a very useful technique for a high thruput analysis of changes in the community structure without clear phylogenetic assignments, which was the aim of this study. However, to link a certain alkB type to the given environmental conditions in a compartment, there is a need to better understand ecophysiology and phylogenetic position of the corresponding organisms. Furthermore, in many cases an analysis of the T-RFLP pattern is done using normalisation; as small peaks are not considered for analysis, this technique may not be able to describe changes in the rare biosphere of alkB harbouring bacteria. As the number of alkB operons and its heterogeneity of so far uncultivated organisms is not known a direct link of number of T-RFs and diversity must be carefully considered. In this respect, sequence-based analysis of metagenomic libraries may help in the future to improve our understanding on alkB harbouring microbes.
Acknowledgments
This work is part of the priority programme 1315 ‘Biogeochemical Interfaces in Soil' and was funded by the German Research Foundation (DFG). Financial support is greatly acknowledged. For support on GC-MS measurement, we are very grateful to Birgit Würz. We wish to thank the three reviewers of the original manuscript for their very helpful and constructive comments, which formed the basis for this revised version.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amouric A, Quéméneur M, Grossi V, Liebgott PP, Auria R, Casalot L. Identification of different alkane hydroxylase systems in Rhodococcus ruber strain SP2B, an hexane-degrading actinomycete. J Appl Microbiol. 2010;108:1903–1916. doi: 10.1111/j.1365-2672.2009.04592.x. [DOI] [PubMed] [Google Scholar]
- Arthurs P, Stiver WH, Zytner RG. Passive volatilization of gasoline from soil. J Soil Contam. 1995;4:123–135. [Google Scholar]
- Avato P, Bianchi G, Pogna N. Chemosystematics of surface lipids from maize and some related species. Phytochemistry. 1990;29:1571–1576. [Google Scholar]
- Bosma TNP, Middeldorp PJM, Schraa G, Zehnder AJB. Mass transfer limitation of biotransformation: quantifying bioavailability. Environ Sci Technol. 1996;31:248–252. [Google Scholar]
- Cayet C, Lichtfouse E. δ13C of plant-derived n-alkanes in soil particle-size fractions. Org Geochem. 2001;32:253–258. [Google Scholar]
- Collister JW, Rieley G, Stern B, Eglinton G, Fry B. Compound-specific δ13C analyses of leaf lipids from plants with differing carbon dioxide metabolisms. Org Geochem. 1994;21:619–627. [Google Scholar]
- Curtis TP, Sloan WT. Exploring microbial diversity—a vast below. Science. 2005;309:1331–1333. doi: 10.1126/science.1118176. [DOI] [PubMed] [Google Scholar]
- de Carvalho CCCR, Wick LY, Heipieper HJ. Cell wall adaptations of planktonic and biofilm Rhodococcus erythropolis cells to growth on C5 to C16 n-alkane hydrocarbons. Appl Microbiol Biotechnol. 2009;82:311–320. doi: 10.1007/s00253-008-1809-3. [DOI] [PubMed] [Google Scholar]
- Dignac M-F, Rumpel C. Relative distributions of phenol dimers and hydroxy acids in a cultivated soil and above ground maize tissue. Org Geochem. 2006;37:1634–1638. [Google Scholar]
- Eglinton G, Gonzalez AG, Hamilton RJ, Raphael RA. Hydrocarbon constituents of the wax coatings of plant leaves: a taxonomic survey. Phytochemistry. 1962;1:89–102. doi: 10.1038/193739a0. [DOI] [PubMed] [Google Scholar]
- Galin T, Gerstl Z, Yaron B. Soil pollution by petroleum products, III. Kerosene stability in soil columns as affected by volatilization. J Contam Hydrol. 1990;5:375–385. [Google Scholar]
- Gans J, Wolinky M, Dunbar J. Microbiology: computational improvements reveal great bacterial diversity and high toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- Gniwotta F, Vogg G, Gartmann V, Carver TLW, Riederer M, Jetter R. What do microbes encounter at the plant surface? Chemical composition of pea leaf cuticular waxes. Plant Physiol. 2005;139:519–530. doi: 10.1104/pp.104.053579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KV, Marschner P, Bünemann EK, Smernik RJ. Chemical changes and phosphorus release during decomposition of pea residues in soil. Soil Biol Biochem. 2007;39:2696–2699. [Google Scholar]
- Hamamura N, Olson SH, Ward DM, Inskeep WP. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl Environ Microbiol. 2006;72:6316–6324. doi: 10.1128/AEM.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider J, Spormann AM, Beller HR, Widdel F. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol Rev. 1999;22:459–473. [Google Scholar]
- Holden PA, LaMontagne MG, Bruce AK, Miller WG, Lindow SE. Assessing the role of Pseudomonas aeruginosa surface-active gene expression in hexadecane biodegradation in sand. Appl Environ Microbiol. 2002;68:2509–2518. doi: 10.1128/AEM.68.5.2509-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar J, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos K, Munch JC, Schloter M. A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J Microbiol Methods. 2006;66:486–496. doi: 10.1016/j.mimet.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–4646. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- Koma D, Hasumi F, Yamamoto E, Ohta T, Chung S-Y, Kubo M. Biodegradation of long-chain n-paraffins from waste oil of car engine by Acinetobacter sp. Soc Biotechnol Jpn. 2001;91:94–96. doi: 10.1263/jbb.91.94. [DOI] [PubMed] [Google Scholar]
- Kotani T, Kawashima Y, Yurimoto H, Kato N, Sakai Y. Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. Soc Biotechnol Jpn. 2006;102:184–192. doi: 10.1263/jbb.102.184. [DOI] [PubMed] [Google Scholar]
- Kotani T, Yurimoto H, Kato N, Sakai Y. Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. Strain TY-5. J Bacteriol. 2007;189:886–893. doi: 10.1128/JB.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranconi MP, Alvarez HM, Studdert CA. A strain isolated from gas oil-contaminated soil displays chemotaxis towards gas oil and hexadecane. Environ Microbiol. 2003;5:1002–1008. doi: 10.1046/j.1462-2920.2003.00507.x. [DOI] [PubMed] [Google Scholar]
- Lichtfouse É, Elbisser B, Balesdent J, Mariotti A, Bardoux G. Isotope and molecular evidence for direct input of maize leaf wax n-alkanes into crop soils. Org Geochem. 1994;22:349–351. [Google Scholar]
- Maffei M. Chemotaxonomic significance of leaf wax n-alkanes in the Umbelliferae, Cruciferae and Leguminosae (Subf. Papilionoideae) Biochem Syst Ecol. 1996;24:531–545. [Google Scholar]
- Manilal VB, Alexander M. Factors affecting the microbial degradation of phenanthrene in soil. Appl Microbiol Biotechnol. 1991;35:401–405. doi: 10.1007/BF00172733. [DOI] [PubMed] [Google Scholar]
- Marin MM, Smits THM, Beilen van JB, Rojo F. The alkane hydroxylase gene of Burkholderia cepacia RR10 is under catabolite repression control. J Bacteriol. 2001;183:4202–4209. doi: 10.1128/JB.183.14.4202-4209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S, Toker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Pérez-de-Mora A, Schulz S, Schloter M.2010MPN- and real time-based PCR methods for the quantification of alkane-monooxygenase homologous genes (alkB) in environmental samplesIn: Cunningham S (ed).Methods in Molecular Biology Human Press: New York; 59–68. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2009R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: Vienna, Austria; ISBN 3-900051-07-0. . http://www.R-project.org . [Google Scholar]
- Richnow HH, Seifert R, Kästner M, Mahro B, Horsfield B, Tiedgen U, et al. Rapid screening of PAH-residues in bioremediated soils. Chemosphere. 1995;31:3991–3999. [Google Scholar]
- Rieley G, Collister JW, Stern B, Eglinton G. Gas chromatography/isotope ration mass spectrometry of leaf wax n-alkanes from plants of differing carbon dioxide metabolisms. Rapid Commun Mass Sp. 1993;7:488–491. [Google Scholar]
- Röling WFM, Milner MG, Jones DM, Fratepietro F, Swannell RP, Daniel F, et al. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with burried oil. Appl Environ Microbiol. 2004;70:2603–2613. doi: 10.1128/AEM.70.5.2603-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepic E, Leskovsek H, Trier C. Aerobic bacterial degradation of selected polyaromatic compounds and n-alkanes found in petroleum. J Chromatogr A. 1995;697:515–523. [Google Scholar]
- Sinowski W, Auerswald K. Using relief parameters in a discriminant analysis to stratify geological areas with different spatial variability of soil properties. Geoderma. 1999;89:113–128. [Google Scholar]
- Smith KEC, Thullner M, Wick LY, Harms H. Sorption to humic acids enhances PAH biodegradation. Environ Sci Technol. 2009;43:7205–7211. doi: 10.1021/es803661s. [DOI] [PubMed] [Google Scholar]
- Top EM, Springael D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol. 2003;14:262–269. doi: 10.1016/s0958-1669(03)00066-1. [DOI] [PubMed] [Google Scholar]
- Totsche KU, Rennert T, Gerzabek MH, Kögel-Knabner I, Smalla K, Spiteller M, et al. Biogeochemical interfaces in soil: the interdisciplinary challenge for soil science. J Plant Nutr Soil Sci. 2010;173:88–99. [Google Scholar]
- Töwe S, Wallisch S, Bannert A, Fischer D, Hai B, Haesler F, et al. Improved protocol for the simultaneous extraction and column-based separation of DNA and RNA from different soil. J Microbiol Methods. 2011;84:406–412. doi: 10.1016/j.mimet.2010.12.028. [DOI] [PubMed] [Google Scholar]
- van Beilen JB, Funhoff EG. Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotec. 2007;74:13–21. doi: 10.1007/s00253-006-0748-0. [DOI] [PubMed] [Google Scholar]
- van Beilen JB, Li Z, Duetz WA, Smits THM, Witholt B. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol. 2003;58:427–440. [Google Scholar]
- van der Meer JR, Sentchilo V. Genomic islands and the evolution of catabolic pathways in bacteria. Curr Opin Biotechnol. 2003;14:248–254. doi: 10.1016/s0958-1669(03)00058-2. [DOI] [PubMed] [Google Scholar]
- Van Hamme JD, Singh A, Ward OP. Recent advances in petroleum microbiology. Microbiol Mol Biol Rev. 2003;67:503–549. doi: 10.1128/MMBR.67.4.503-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez S, Nogales B, Ruberto L, Hernández E, Christie-Oleza J, Lo Balbo A, et al. Bacterial community dynamics during bioremediation of diesel oil-contaminated Antarctic soil. Microb Ecol. 2009;57:598–610. doi: 10.1007/s00248-008-9420-9. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Baldock JA, Dalal RC, Moody PW. Decomposition of plant materials in relation to nitrogen availability and biochemistry determined by NMR and wet-chemical analysis. Soil Biol Biochem. 2004;36:2045–2058. [Google Scholar]
- Whyte LG, Hawari J, Zhou E, Bourbonnière L, Inniss WE, Greer CW. Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl Environ Microbiol. 1998;64:2578–2584. doi: 10.1128/aem.64.7.2578-2584.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick LY, Colangelo T, Harms H. Kinetics of mass transfer-limited bacterial growth on solid PAHs. Environ Sci Technol. 2000;35:354–361. doi: 10.1021/es001384w. [DOI] [PubMed] [Google Scholar]
- Widdel F, Rabus R. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol. 2001;12:259–276. doi: 10.1016/s0958-1669(00)00209-3. [DOI] [PubMed] [Google Scholar]
- Wiesenberg GLB, Schwarzbauer J, Schmidt MWI, Schwark L. Source and turnover of organic matter in agricultural soils derived from n-alkane/n-carboxylic acid compositions and C-isotope signatures. Org Geochem. 2004;35:1371–1393. [Google Scholar]
- Wilkes H, Kühner S, Bolm C, Fischer T, Classen A, Widdel F, et al. Formation of n-alkane- and cycloalkane-derived organic acids during anaerobic growth of a denitrifying bacterium with crude oil. Org Geochem. 2003;34:1313–1323. [Google Scholar]
- Yaalon DH. Down to earth. Nature. 2000;407:301. doi: 10.1038/35030260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.