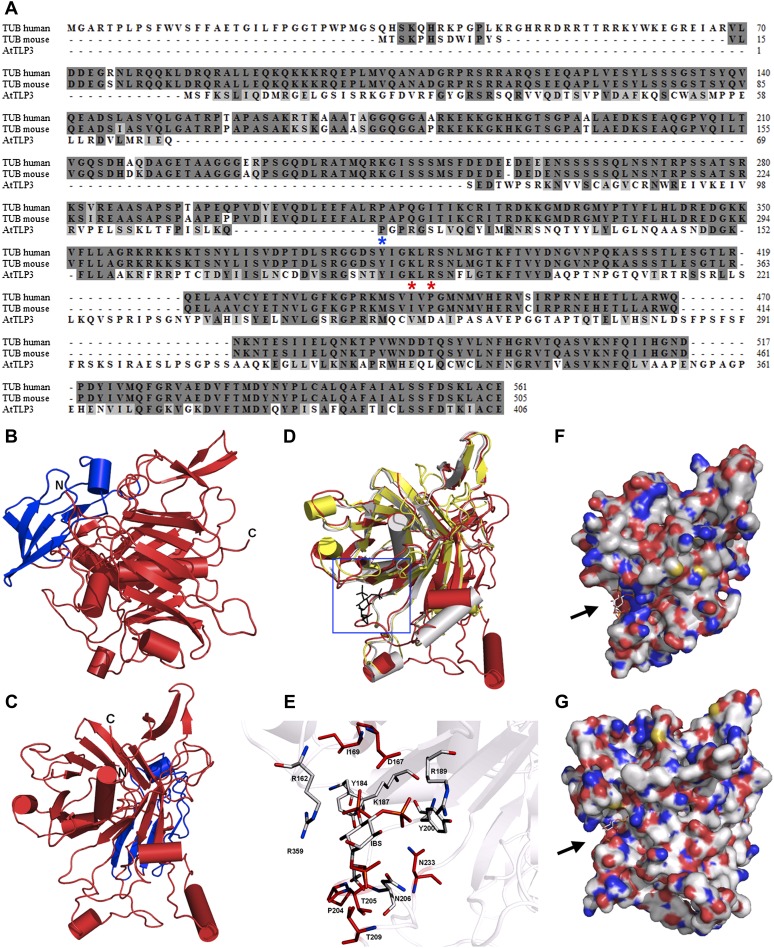
Figure 2.
Sequence and 3D structure of AtTLP3 and its Tubby domain. A, Sequence alignment for human (accession no. 19923167) and mouse (accession no. 11230782) Tubby (TUB) protein with AtTLP3 (accession no. 30690823). Identical and similar amino acids are shaded in dark gray and light gray, respectively. The two amino acids that are necessary for PIP2 binding (Santagata et al., 2001) are indicated (red asterisks) underneath those amino acids. The blue asterisk underneath one amino acid indicates the start of the Tubby domain. B and C, 3D homology model of AtTLP3 showing the FB domain (blue) and the Tubby domain (red). The N and C termini are highlighted. D, Superposition of the structural model of AtTLP3 (red) with the crystal structure of the human Tubby protein (PDB identifier 1S31; yellow) and the crystal structure of mouse brain Tubby protein (gray) bound to PIP2 (black, blue frame). E, Magnification of the frame in D shows the highly conserved inositol lipid-binding domain of AtTLP3. A comparison of AtTLP3, mouse, and human Tubby proteins was done. The PIP2 analog l-α-glycerophospho-d-myoinositol 4,5-bisphosphate (IBS) was used to show the conservation of binding sites. Amino acid residues that are similar in type and position (conformation) in all three proteins are colored in gray, and amino acids that are specific for the AtTLP3 protein are marked in red. F and G, Surface representation of the mouse Tubby protein with bound IBS (F) and structural model of AtTLP3 (G) with modeled IBS. The groove of highly positive charge (blue regions) is more significant in the mouse protein, and the dimension of the groove differs in the AtTLP3 structural model.