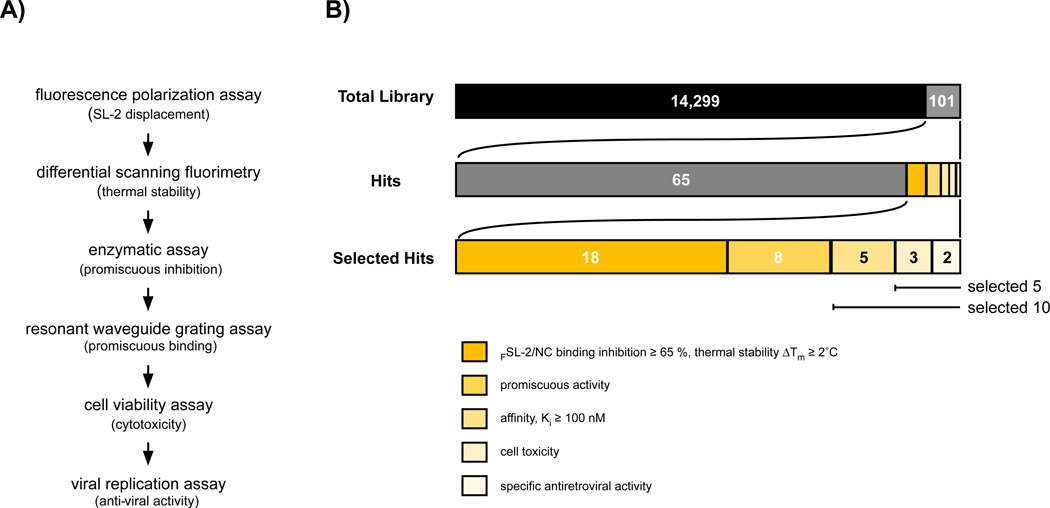
Figure 2. Overview of the strategy for identifying compounds that disrupt HIV-1 nucleocapsid/RNA interaction.
(A) Outline of the experimental procedure. (B) Enumeration of Hits during library screening. Utilizing a fluorescence polarization assay to detect small molecule displacement of 3'-6-carboxyfluorescein-labeled SL-2 DNA (FSL-2) from binding to the nucleocapsid (NC’) protein, 14,400 compounds were screened and 101 compounds (Hits) were identified as displacing FSL-2. The 101 Hits were retested for NC’-FSL-2 displacement and 65 did not show activity or demonstrated < 60 % FSL-2 displacement activity and were eliminated from further study. Of the remaining 36 compounds (Selected Hits), 18 failed to show a significant shift in thermal stability when the compound-Gag complexes were evaluated by differential scanning fluorometry assay, 8 displayed promiscuous β-galactosidase inhibition, and 5 were excluded due to low affinity binding to NC’. Of the 5 remaining Selected Hits, 3 Hits showed cellular toxicity at higher compound concentrations, whereas, 2 compounds demonstrated both low cellular toxicity and inhibition of HIV-1 replication in CD4+ T cells.