Figure 2.
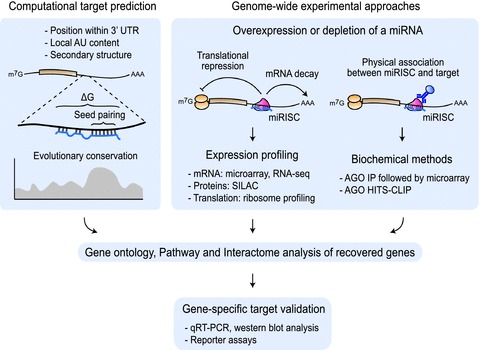
Genome-wide probing for miRNA targets. Computational algorithms for miRNA target prediction rely on a set of established features of miRNA–target interactions, including seed pairing, free energy of the miRNA-target heteroduplex, local AU content, secondary structure, and evolutionary conservation. Genome-wide experimental approaches often start with differential expression of a particular miRNA by overexpression or knockdown. Gene sets for which mRNA or protein levels respond to the perturbation are identified by various expression profiling methods. Alternatively, putative target mRNAs can be directly enriched by Ago immunoprecipitation. Gene ontology and interactome analysis of recovered genes might help elucidate the biological functions of the miRNA. Finally, individual miRNA–target interactions are validated by reporter assays.
