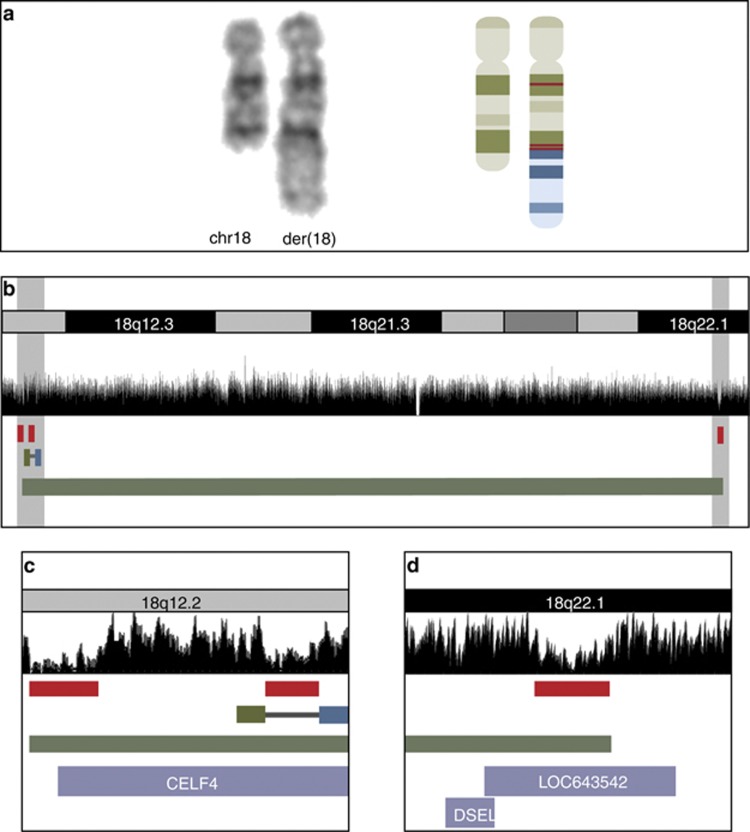
Figure 2.
Partial karyotype and molecular findings on the derivative chromosome 18. (a) A partial karyotype showing the normal and the derivative chromosome 18, and a schematic representation of the two chromosomes and the identified rearrangements; the red lines represent deletions associated with the translocation breakpoint at 18q12.2 and the inversion breakpoints at 18q12.2 and 18q22.1. (b) The breakpoint region at 18q. The mate pair coverage is shown in black. Deletions detected by SNP 6.0 array is represented by red bars; note the corresponding low coverage of the mate pair reads. The missing coverage of mate pair reads in 18q21.3 corresponds to a gap in the annotated reference genome (UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly). The translocation breakpoint is represented by green reads mapping to 18q12.2 and blue reads mapping to 12q21.31, and the inversion is represented by a green bar. (c, d) Zoom in on the breakpoint regions (gray shaded areas in b). The protein coding gene CELF4 is truncated by the translocation and two deletions at 18q12.2 (c), and the non-coding RNA LOC643542 is truncated by the inversion breakpoint and an associated deletion at 18q22.1 (d).