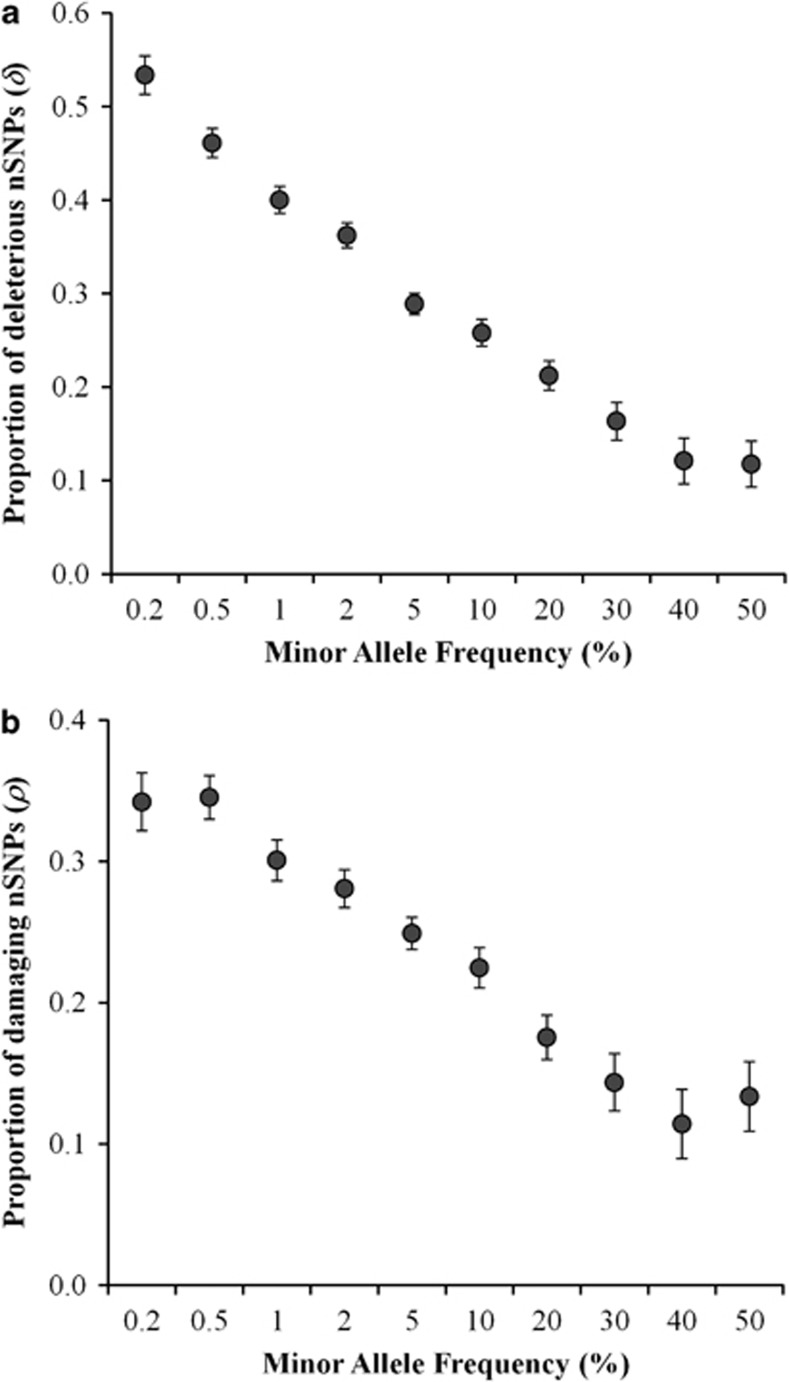
Figure 1.
(a) Deleterious fraction of nSNPs (δ) estimated for variants with different minor allele frequencies (MAF). SNPs were grouped into 10 categories based on their MAF and only the upper values of the ranges are shown (X-axis). Error bars denote the SE, which was estimated using a bootstrap procedure (1000 replications). (b) The proportion of damaging nSNPs (ρ) was estimated using amino acid variants belonging to 10 MAF categories. Error bars indicate the SE computed using the binomial variance.