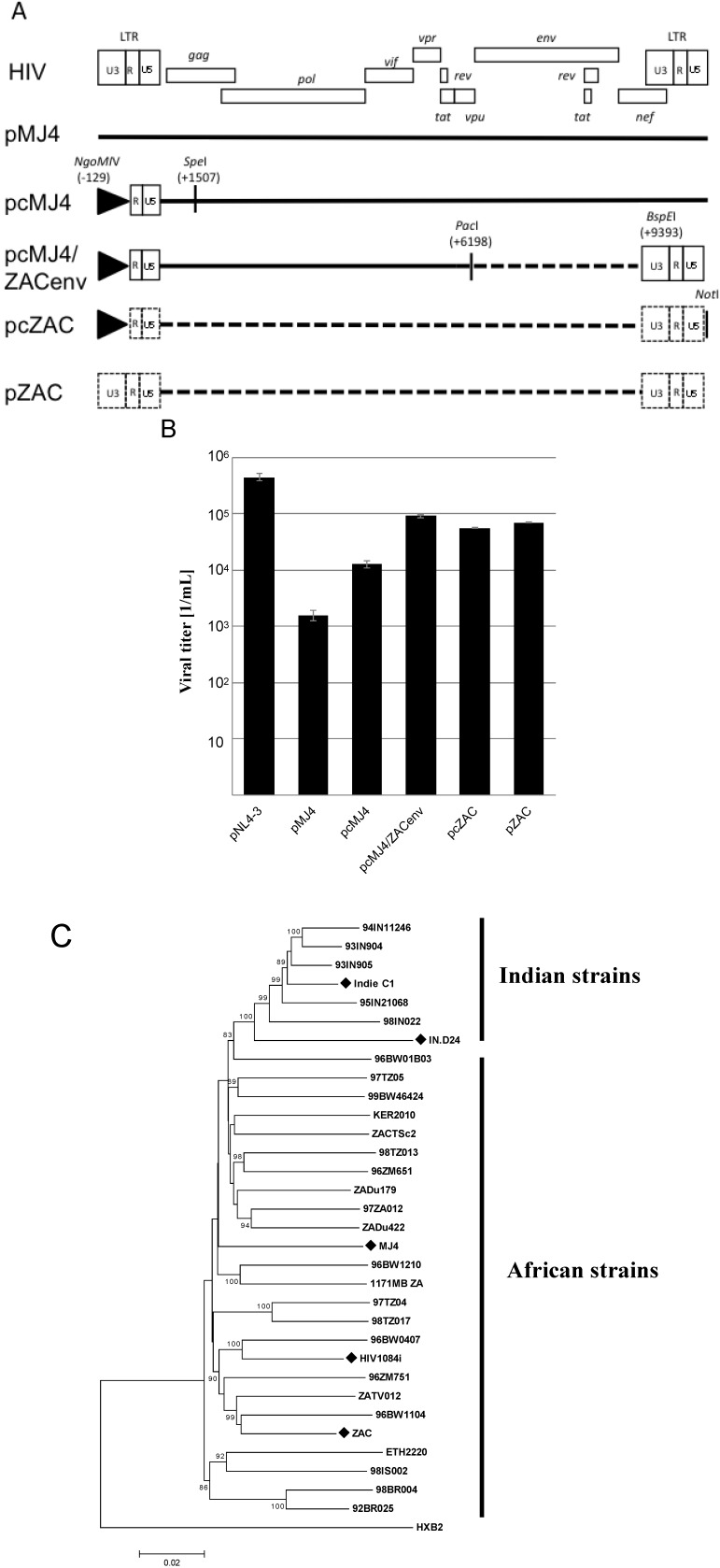
Figure 1.
Molecular characterization of pZAC, an infectious proviral clone from Cape Town, South Africa. (A) Cloning strategy used during the study. The U3 promoter of pMJ4 was replaced with a CMV-IE promoter, as indicated. The CMV-promoter is represented by a triangle. The proviral clone pcMJ4 was used as a backbone for further characterization of pZAC. Dotted lines indicate patient ZAC derived sequences. The enzyme restriction sites used for cloning are indicated.(B) Transient virus titer on TZM-bl of infectious proviral clones. After transfection of HEK 293T cells, cultured supernatants were titrated on HeLa TZM-bl indicator cells to determine the transient viral titer of HIV proviral clones [16]. Titers and standard deviation were derived from three independent experiments. (C) Phylogenetic analysis of HIV-1 subtype C infectious clones. A Neighbour-Joining tree was drawn from the infectious HIV-1 subtype C clones, compared to a HIV-1 subtype C reference set (dataset obtained from [17]). Evolutionary distances were calculated using the Maximum Composite Likelihood method, with a bootstrap test of 10,000 replicates. The branch scale, indicating the evolutionary distance, is indicated. The Indian and African strains form two unique phylogenetic clusters, with the newly described pZAC sequence showing similarity to the Botswana HIV-1 subtype C sequences, 96BW1104. The HIV-1 subtype B reference sequence, HXB2 was used as an outlier to root the phylogenetic tree.