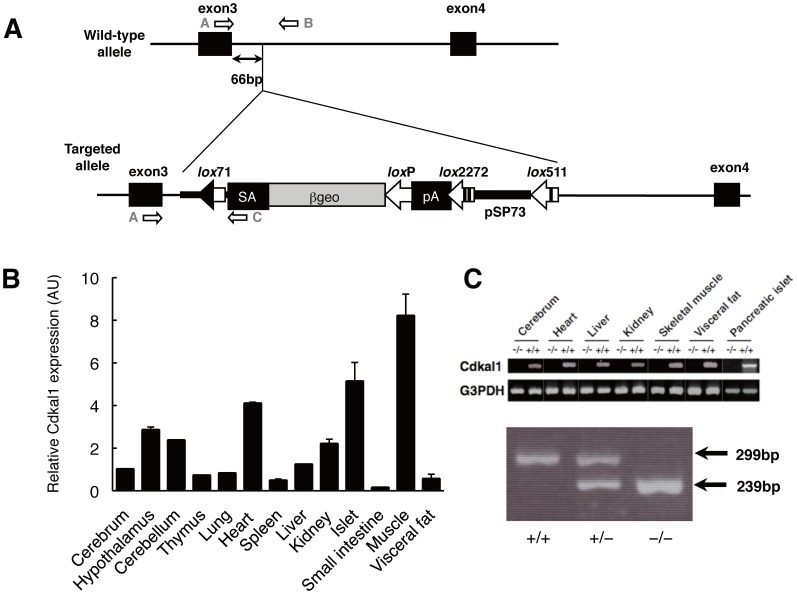
Figure 1. Generation of Cdkal1-knockout mice.
A: Integration site of the trap vector. Filled boxes represent exons 3 and 4 of the Cdkal1 gene. The trap vector was inserted 66-bp downstream from exon 3. The trap vector pU17 contains an intron and a splice acceptor (SA) sequence from the mouse En-2 gene, the ßgeo gene, and polyadenylation signal (pA). A lox71 site is located within the intron sequences, and loxP, lox2272, and lox511 sites are located downstream of the ßgeo, pA, and pSP73 vector sequences, respectively. The arrows indicate the primers used for genotyping. The primer sequences were: (A) CGAAGGCGGAATACCCAAA, (B) TCATATCCGTTCCCCTAATTCCC, and (C) GTCCCCCTTCCTATGTAACCCAC. B: Relative mRNA expression of Cdkal1 in a series of tissues/organs from C57BL/6 mice. Total RNA was isolated from various tissues of mice, treated with DNase, and then reverse-transcribed using ReverTra Ace (Toyobo, Osaka, Japan) and random primers. PCR was performed using a primer pair to specifically amplify part of the mouse Cdkal1 gene (1090 bp in size), from exon 3 to 12∶5′-CGAAGGCGGAATACCCAAA-3′ and 5′-CAGCAGGAGTTCCGGGTCTT-3′. Quantitative reverse-transcription (RT) PCR was performed using a Cdkal1-specific primer and probe (Mm00507443_m1) (Applied Biosystems, Foster City, CA, USA). The values were arbitrary units after normalization with actin, beta (Actb). C: RT-PCR analysis of selected tissues of Cdkal1 +/+and Cdkal1 −/− mice (upper panel). Gapdh mRNA was used as a positive control. PCR analysis for genotyping (lower panel). DNA fragments from the wild-type (299 bp) and target (239 bp) alleles were amplified using primer pairs A/B and A/C, respectively.