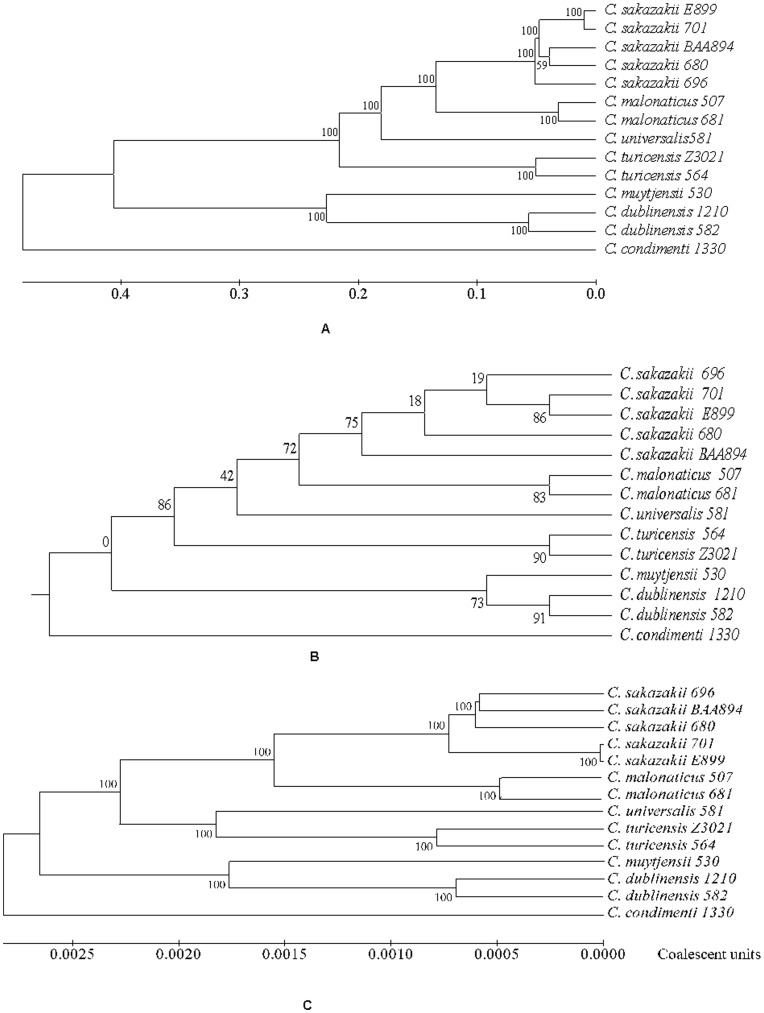
Figure 1. Phylogentic analysis of Cronobacter spp. based on whole genome sequencing.
(A) A maximum likelihood phylogram based on 138,143 SNPs identified from a concatenated alignment of 1117 core genes (879,768 nucleotides) present in single copies in all strains. The numbers on the nodes represent the bootstrap support from 1000 replicates (in percentage). (B) A majority rule consensus tree generated by summarizing individual 1117 core gene trees. The numbers on the internal nodes indicate the fraction (in percentage) of gene trees which support the partition of the taxa into the two sets. (C) A Clonal Frame (CF) analysis based on randomly selected 99 core loci. The consensus tree was obtained from 2 CF runs with 200,000 MCMC iterations. The numbers on the nodes represent the bootstrap support in percentage.