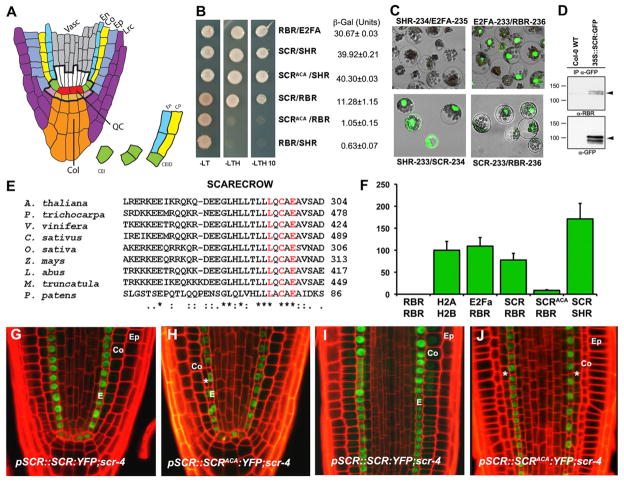
Figure 1. A Conserved LxCxE Motif in SCR Mediates Direct Binding to RBR.
(A) Root stem cell niche organization and cell transitions and divisions defining ground tissue lineages.
(B) Yeast two-hybrid analyses showing and quantifying SCR-RBR interaction. RBR-E2FA and SCR-SHR combinations are positive controls, and RBR-SHR is the negative control.
(C) SCR-RBR binding by BiFC in Arabidopsis mesophyll protoplasts. RBR-E2FA and SCR-SHR are positive controls.
(D) Coimmunoprecipitation of RBR with α-GFP antibody in WT and 35S::SCR:GFP root extracts. Black arrow marks endogenous RBR in top panel and SCR-GFP in lower panel.
(E) Protein sequence alignment of SCR orthologs in seed plants and P. patens moss showing conservation of the LxCxE motif.
(F) In vivo interaction strengths from split Renilla luciferase assay in mesophyll protoplasts. RLUs were normalized to H2A-H2B interaction strength. Arrow bar represents SEM.
(G–J) Confocal laser scanning microscope (CLSM) of longitudinal root sections of 5 dpg. scr4-1 plants complemented with WT SCR (G and I) and SCRAxCxA (H and J). Ep, epidermis; Co, cortex; E, endodermis;*, extra ground tissue layer. See also Figure S1.