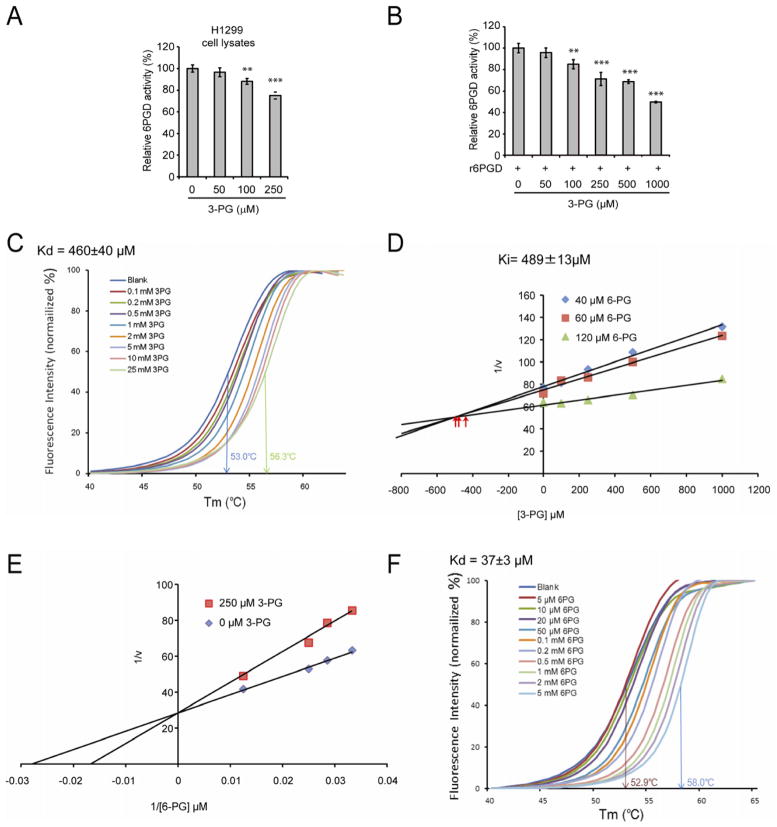
Figure 2. Attenuation of PGAM1 results in increased intracellular levels of 3-PG, which binds to and inhibits 6PGD by competing with its substrate 6-PG.
(A–B) Enzyme activity of 6PGD in H1299 cell lysates (A) or recombinant 6PGD (r6PGD) (B) was determined in the presence of increasing concentrations of 3-PG. Relative 6PGD activity was normalized to the control samples without 3-PG treatment. 3-PG levels in control H1299 cells with empty vector and PGAM1 knockdown are 62.5±10.8μM and 256±41.9μM, respectively. The error bars represent mean values +/− SD from three replicates of each sample (**: 0.01<p<0.01; ***: p<0.001).
(C) Thermal shift melting curves of 6PGD and 3PG. Thermal shift assay was performed to examine the protein (6PGD) and “ligand” (3PG) interaction. Change of melting temperature (Tm) in a dose-dependent manner at concentrations from 100 μM to 25 mM demonstrates that 3-PG directly binds to the protein. Kd for 6PGD-3-PG interaction was determined to be 460±40 μM.
(D) The Dixon plot shows that 3-PG inhibits 6PGD and the dissociation constant (Ki) was determined.
(E) The Lineweaver-Burk plot shows that 3-PG functions as a competitive inhibitor of 6PGD.
(F) Thermal shift melting curves of 6PGD and 6PG. Thermal shift assay was performed to examine the protein (6PGD) and ligand (6PG) interaction. Change of melting temperature (Tm) in a dose-dependent manner at concentrations from 5 μM to 5 mM demonstrates that 6-PG directly binds to the protein. Kd for 6PGD-6PG interaction was determined to be 37±3μM.
See also Figure S2.