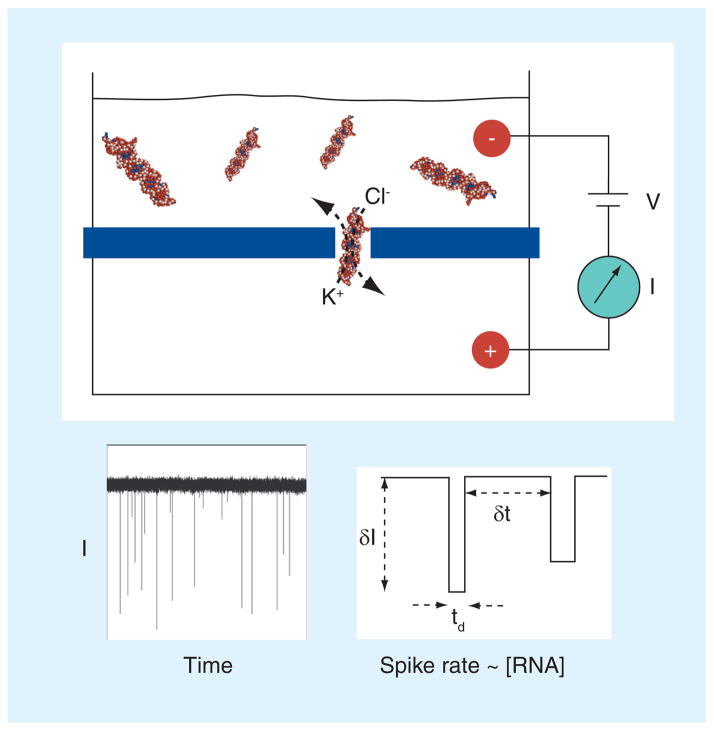
Figure 1. Scheme of nanopore-based single-molecule detection.
Voltage is applied using a pair of electrodes that provide an electrochemical circuit with the solution. The voltage (V) drives ions through the nanopore, which results in a measurable steady-state ‘open pore’ current (I). When a biomolecule in the top chamber diffuses into the pore, it blocks the ion pathway of the pore and the flux of ions is reduced, resulting in a negative spike that signals each molecule. Under the same applied voltage conditions, the rate of spike events has a linear correspondence with the macromolecular concentration. Therefore, miRNA duplexes can be counted and quantified by measuring their rate of passage through the pore. A real current trace is shown for illustration, as well as three parameters that are typically quantified in nanopore experiments. δI and td are used to identify the target blocks, while δt is used to calculate the rate of block occurrence, which is the inverted value of δt.
δI: Current block amplitude; δt: Interval between adjacent blocks; I: Current; td: Block duration; V: Voltage.