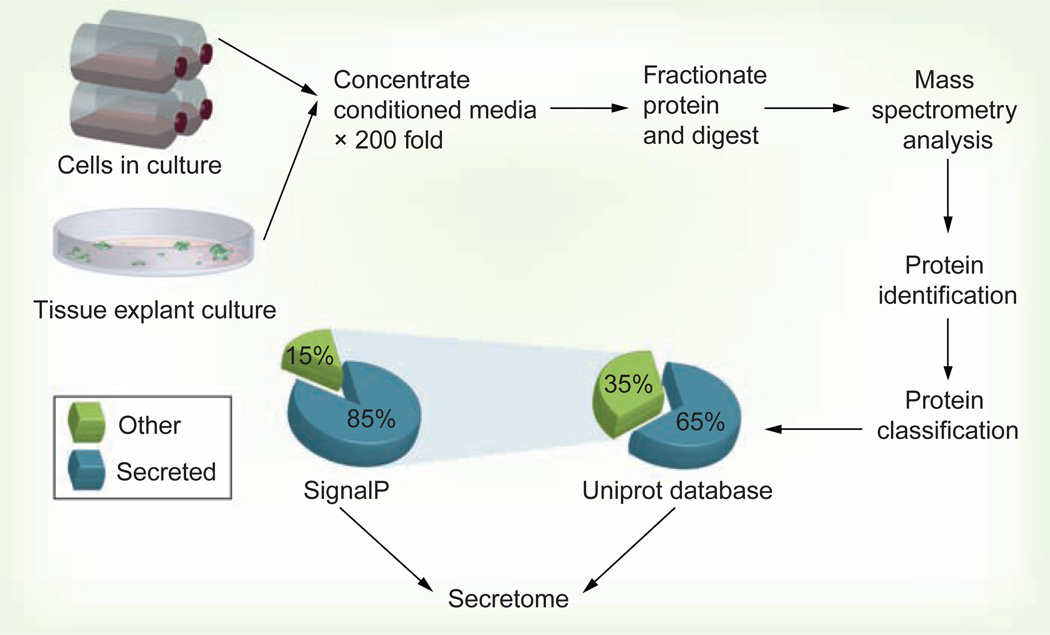
Figure 1. Generic in vitro secretome profiling flow chart.
Typically, cells are grown to 80 or 100% confluence in serum-supplemented media, then washed (up to six times with serum-free medium or phosphate-buffered saline) and incubated for an additional 12–24 h in serum-free media. Conditioned medium containing secreted proteins is collected, centrifuged at 300 g to remove floating cells and gross cell debris, then further passed through a 0.22-µm filter to remove small debris. Conditioned medium is then concentrated up to 200-fold using a centrifugal filtration device (usually with a 3-kDa molecular weight cutoff). Protein concentration is measured and usually 50–100 µg of total protein is fractionated by SDS-PAGE. The resulting lane is sliced into 30–35 bands for in-gel digestion and LC-MS/MS analysis. Resulting data are searched against the desired protein database using a suitable search engine (such as Sequest or Mascot). The obtained list is then further curated for protein subcellular localization using the Uniprot knowledgebase [104]. Unclassified proteins are checked for potential secretion using the SignalP server [105]. Other proteins can be searched manually for subcellular localization in the literature or exosome databases.