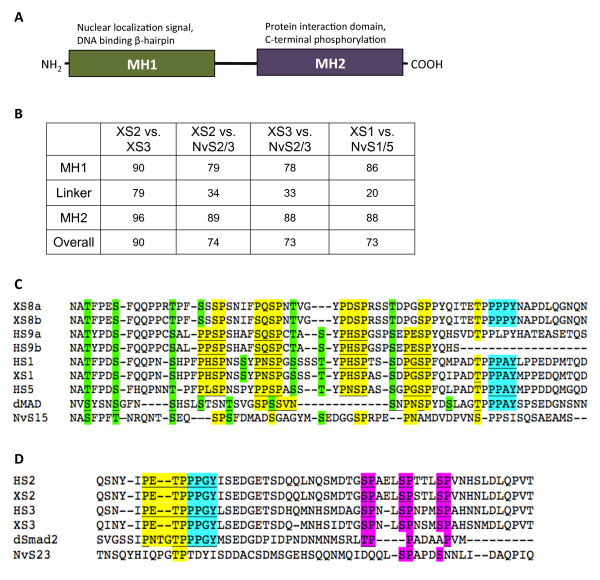
Figure 1.
R-Smads are defined by two conserved protein domains. (A) Diagram of a typical R-Smad showing the two conserved domains (MH1 and MH2) with important regions noted. (B) A table of pairwise percent identity of the MAD homology domains and the non-conserved linker regions of Xenopus and Nematostella R-Smads [see Additional file 1 for a detailed amino acid alignment and Additional file 2 for a table of GenBank accession numbers]. (C) Alignment of relevant sections of the Smad1/5/8 linker regions from multiple taxa. The Smurf1 site PPXY motif is highlighted in cyan, MAPK recognition sites are highlighted in yellow, and GSK3 consensus sites are highlighted in green. (D) Alignment of relevant sections of the Smad2/3 linker regions from multiple taxa. The Smurf2 site PPXY motif is highlighted in cyan, MAPK recognition sites are highlighted in yellow, and proline-directed kinase sites are highlighted in magenta. Underlined sequences in (C) and (D) indicate consensus sites identified for the particular protein and species in the literature; all other highlights reflect our inferences based on the alignments.