Figure 1.
Activity-Dependent Changes in GluA1 and GluA2 RNA Processing Localized to Hippocampal CA1
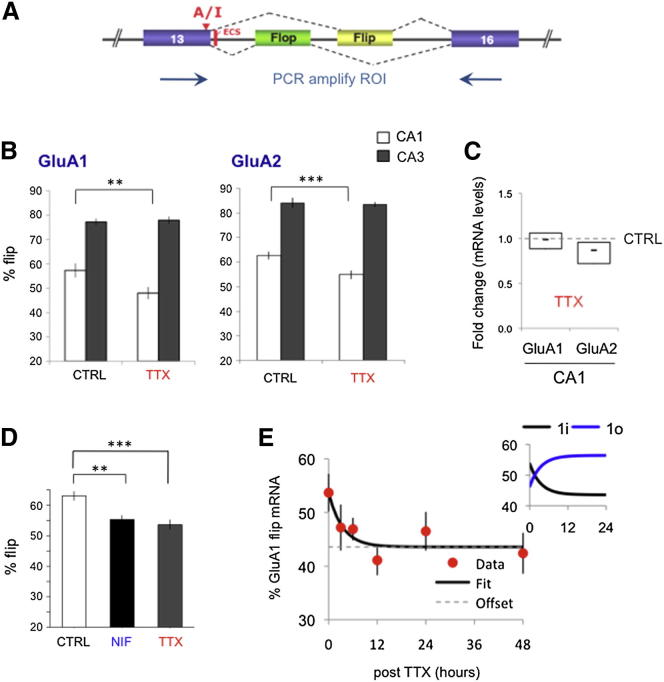
(A) Schematic of gria2 exons 13–16; the region of interest (ROI) amplified from cDNA encodes the i/o exons and the A/I (adenosine-to-inosine) RNA editing site. The editing complementary sequence (ECS) forms a conserved pre-mRNA secondary structure encompassing the splice site. (B) Quantification of peak heights in CA1 sequence chromatograms. Plots show the abundance of flip splice variants for A1 and A2 as a fraction of total subunit mRNA for ±TTX. Quantification of splice variants is determined from mean peak height ratios for the first five alternatively spliced nucleotides. Two-tailed t test, ∗∗p < 0.005; ∗∗∗p < 0.0001. (C) TaqMan real-time PCR measurements (A1 or A2/Gapdh) reveal no detectable differences between A1 and A2 mRNA expression after TTX relative to CTRL (dashed line). (D) Reduction of A2 flip levels in response to chronic nifedipine and to chronic TTX. ANOVA, p < 0.001; Dunnett’s multiple comparison (∗∗CTRL versus NIF; ∗∗∗CTRL versus TTX). (E) Time course of i/o splicing changes in response to TTX. Slices were harvested 0, 3, 6, 12, 24, and 48 hr post-TTX. The sample size was 9–17 slices/time point. The amount of A1i as a percentage of total A1i/o was determined from peak measurements of sequence traces. Data points were fit with a single exponential (y = Ae–τ/t + c). Since alternative splicing of i/o exons is mutually exclusive and overall A1 levels remain unchanged (C), a decrease in A1i is accompanied by an increase in A1o mRNA (inset).