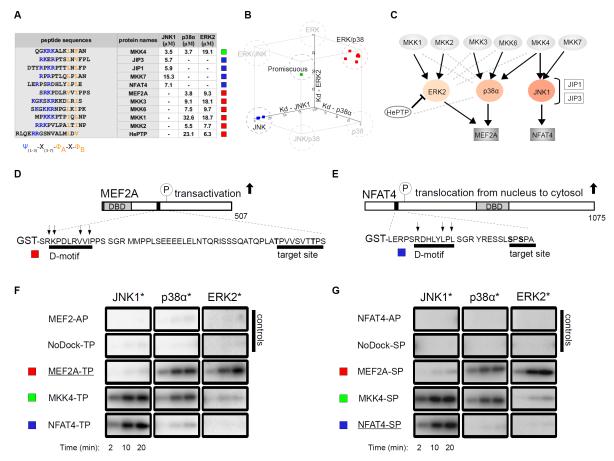
Figure 1. Biochemical specificity of classical D-motifs.
(A) Binding affinities of chemically synthesized D-motif peptides with JNK1, p38α and ERK2. “-” indicates no detectable binding, Kd > 100 μM; N=3 experiments. (B) These binding affinities are plotted on a three-dimensional scatter plot in which axes represent dissociation constants (Kd) for the three different MAPKs and squares correspond to individual peptides listed in the table on the left. (C) The in vivo wiring diagram for MAPKs with their partners. Black connections indicate physiologically relevant links; gray dashed lines indicate binding that does not concur to physiologically relevant connections. (D-G) Docking motifs govern the phosphorylation of critical transcription factor regions in MEF2A (D) and NFAT4 (E) by MAPKs. A representative set of phosphor imaging results of SDS-PAGE gels are shown from at least two in vitro kinase experiments using activated (*) MAPKs to phosphorylate control, “wild-type” or D-motif MEF2A (F) and NFAT4 (G) chimera reporters. AP constructs: Thr or Ser residues in target S/TP site were mutated to alanines; NoDock constructs: the basic and ΦxΦ motif residues, indicated by arrows, were mutated to alanines; DBD, DNA-binding domain.