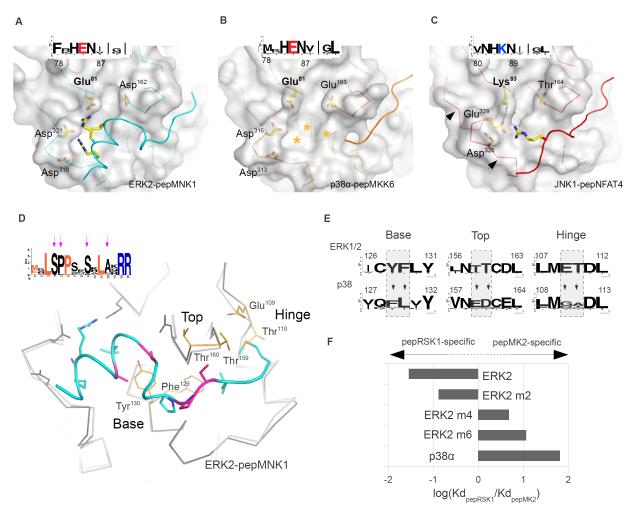
Figure 4. Topography of paralogous MAPK docking grooves.
(A-C) Structural basis for the ability of JNK to discriminate between certain D-motifs and RevD-motifsPeptide-bound docking surfaces of ERK2 (A), p38α (B) and JNK1 (C) are presented in the same orientation. Arrows on (C) indicate the critical region in JNK that determines the width of the CD groove, which in turn also influences the size of the upper pocket. The insets show the sequence logo of the corresponding regions in MAPK homologs from sponges to human. Stars denote the tentative position of the pepMKK6 basic region in the p38α CD groove. (D) Structural determinants of p38-ERK discrimination. Arrows indicate conserved amino acids in the pepMNK1 sequence logo that are not part of the revD-motif consensus, which are colored in magenta on the human MAPK:revD-motif complex structure below. Superimposed p38α is colored in light gray and shown as a Cα-trace. (E) The evolutionarily conserved nature of the base, top and hinge is shown for ERK1/2 and p38 homologs from sponges to human from the KinBase database (73) as sequence logos. (F) The impact of amino acid swaps in the ERK2 docking groove on pepRSK1 or pepMK2 binding is shown as a bar diagram. Amino acid swaps were done in the base (m2), in the base and the top (m4) and in all three regions (m6) of ERK2. New structures shown on this figure are p38α-pepMKK6, JNK1-pepNFAT4 and ERK2-pepMNK1.