Figure 1.
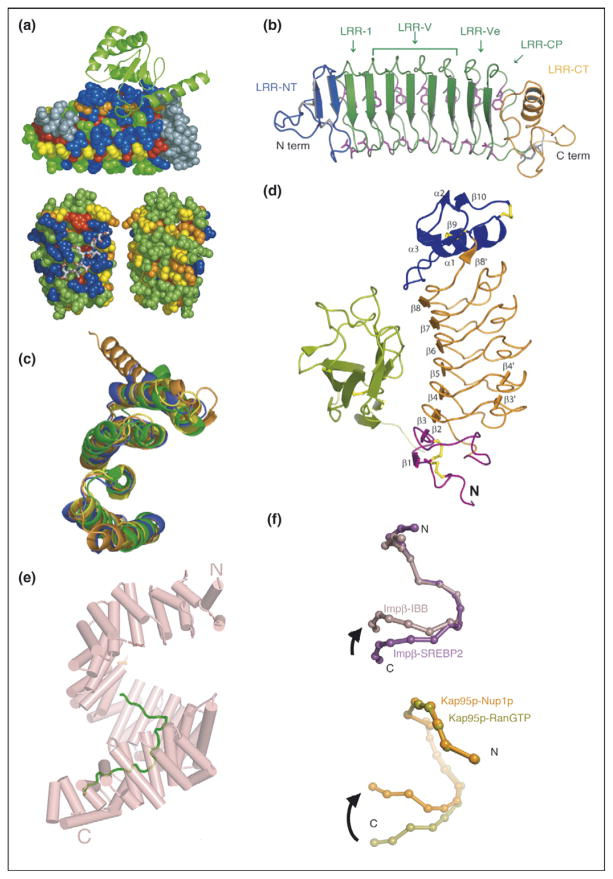
Repeat proteins as conserved scaffolds. (a) Sequence variability analysis repeat proteins. Upper panel: variability analysis of 15 497 Ank repeats. The 4-Ank domain from GABPβ1 (spheres) is shown bound to the ligand GABPα (ribbons) (PBD ID: 1awc). Coloring is from most variable = blue to least variable = red. The binding surface is clearly more blue than the rest of the proteins’ surface. Lower panel: sequence variability analysis of 6887 TPR repeats mapped onto the co-crystal structures of HOP-TPR1/Hsc70 peptide (PBD ID: 1elw) with the TPR domain rendered in spheres and the ligands in sticks. Two views from 180° rotation of the molecule are shown. The concave, ligand binding surface, left, is clearly more variable (blue) than the convex, solvent exposed surfaces (green, yellow), right [1,3]. (b) Crystal structure of hagfish variable lymphocyte receptor (VLR-A). The VLR-A protein comprises 8 LLR modules. The constant N-terminal (LRR-NT) and C-terminal (LRR-CT) modules are shown in blue and orange, respectively. The variable LRR modules (LRR-V) that can be present in variable numbers are colored in green [2••]. (c) Alignment of TPR domains. Structural alignment of TPR domains unliganded and peptide-bound: CTPR3 (PDB ID 1NA0) [35] in blue, TPR domain (residues 19-177) of PP5 in orange (PDB ID: 1a17) [36], and TPR2A (PDB ID: 1elr) in green and TPR1 (PDB ID: 1elw) in yellow, domains of Hop from the co-crystal structures with their peptide ligands, C-terminal peptides of Hsp90 and Hsp70, respectively [37]. The structures are all completely superimposable, with backbone RMSD values that vary from 1.1 to 1.9 Å for different pairwise structural alignments [5]. (d) Structure of Slit2 D2 bound to Robo1 Ig1. Ig1 is in green; Slit2 D2 N-terminal and C-terminal caps are in purple and blue, respectively; LRRs 1–6 are in orange; and the disulfide bridges are in yellow [6]. (e) Structure of HEAT domain of karyopherin Kapβ2 bound to NLS substrate (PDB ID: 1ot8). The helices are represented as cylinders and structurally disordered loops as dashed lines. The extended NLS sequence of the cargo protein hnRNPA1 is shown in green. A1-NLS peptide binds in an extended conformation on the concave surface of the C-terminal arch of the karyopherin superhelix [10••]. (f) Schematic illustration of the hinge motion between subgroups of HEAT repeats. (Top) Schematic illustration of the hinge movement between HEAT repeat subdomains H1–H12 and H14–H19 in Impβ and Kap95p upon ligand binding. IMM-bound Impβ (pink) and SREBP2-bound Impβ (purple) are superimposed at HEAT repeats 5–10, with a sphere representing the center of each HEAT repeat. (Bottom) Schematic illustration of the hinge movement between HEAT repeat subdomains H1–H14 and H15–H19. RanGTP-bound Kap95p (light green) and Nup1p-bound Kap95p (orange) are superimposed at HEAT repeats 6–13, with a sphere representing the center of each HEAT repeat [10••].