FIGURE 1.
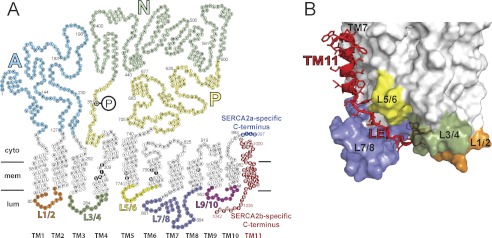
Amino acid sequence and membrane topology of SERCA isoforms 2a and 2b, illustrating the difference in the C-terminal tail region (A), and three-dimensional model of the structural relationship of the extended C terminus of SERCA2b (B). In A, each circle corresponds to an amino acid residue indicated by the single-letter code inside the circle. The phosphorylated aspartate (Asp351) is indicated by white lettering on black background with a large encircled P attached. TM1-TM11 denotes the transmembrane helices, of which TM11 is found only in SERCA2b. In the transmembrane helices, circles with white lettering on black background indicate the calcium binding residues. The unique C-terminal tail of SERCA2b is colored red, whereas the corresponding part of SERCA2a is shown in blue. SERCA2a and SERCA2b are identical in the first 993 amino acid residues. SERCA1a (994 residues, not shown) has a topology similar to SERCA2a with 84% amino acid identity. The SERCA2b-specific tail comprises in addition to TM11 also an 11-amino acid luminal extension (residues 1032–1042). The cytoplasmic domains A, N, and P, are indicated using different color codes, as are the luminal loops L1/2, L3/4, L5/6, L7/8, and L9/10. B, the C-terminal tail of SERCA2b modeled on a SERCA2a homology model derived from the crystal structure of SERCA1a in Ca2E1 form (6). The modeled TM11 is in proximity to TM7 and TM10. As it descends from TM11 the luminal extension is in close contact with the luminal loop L7/8. The last four residues, MFWS, occupy a luminal docking site formed by luminal loops L1/2, L3/4, and L7/8 (6). The luminal loops are indicated by the same color code as in A.
