Fig. 3.
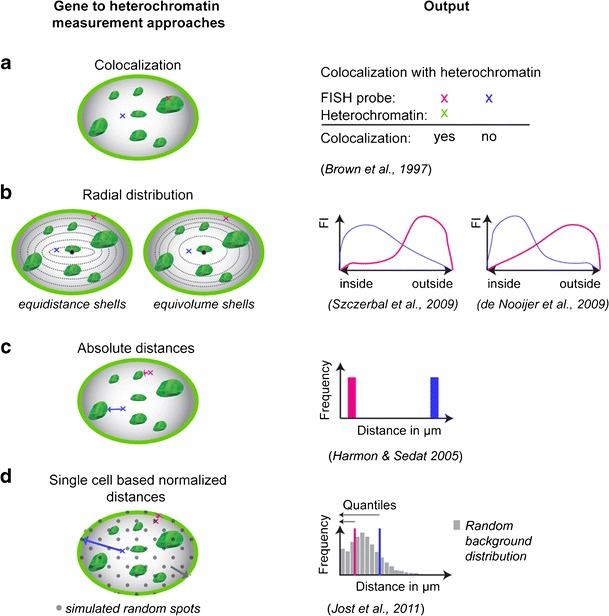
Strategies to investigate the role of heterochromatin (proximity) in gene silencing. The schemes (left) present alternative approaches to determine gene (red and blue) to heterochromatin (green) proximity in the nucleus and their respective data outcome (right). a Classic evaluation is based on simple colocalization of genes and heterochromatin yielding a clear yes or no outcome. b Radial distribution analysis within the cell nucleus can be performed in shells of the same distance (equidistance) or of the same volume (equivolume). Both methods yield a fluorescence intensity (FI) based distribution. Depending on which shell distribution is chosen, either interior or exterior genes are underestimated. c Comparing gene to heterochromatin absolute distances in 3D is a very unbiased method but does not take into account morphological variability. d The quantile-based approach combines absolute distance measurements (in 3D) with single-cell normalization (software and audiovisual tutorial available at http://www.cardoso-lab.org/publications/Randomizer.zip). Simulation and measurement of random (virtual) spots (gray) results in a random background distribution that normalizes for morphological differences. For each procedure and outcome, references are indicated below. An annotated list of studies is given in Table 1
