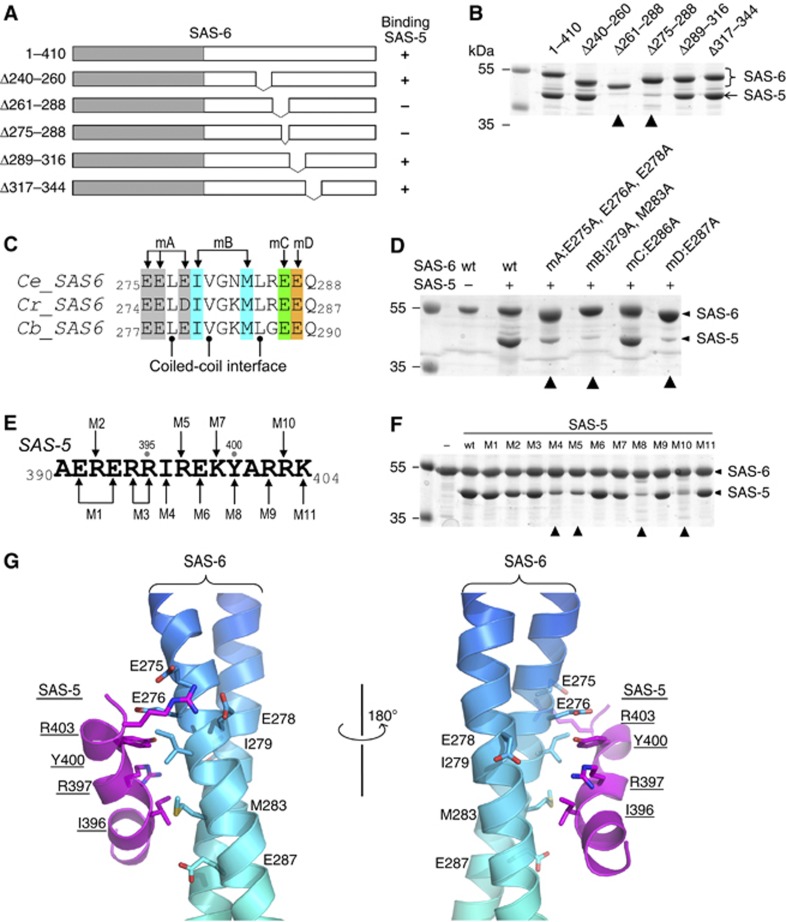
Figure 3.
Association of the SAS-5 CTD and the SAS-6 CCD is mediated by synergistic hydrophobic and electrostatic interactions. (A) Schematic of SAS-6 deletion constructs. The right column summarizes the interaction results in (B). (B) In vitro pull-down results of MBP-tagged SAS-5 CTD using Ni-NTA bound SAS-6 as the bait. The two deletions of SAS-6 that failed to pull down SAS-5 are indicated with arrowheads. (C) Sequence alignment of the SAS-5-binding site from three Caenorhabditis species. Ce, Caenorhabditis elegans; Cr, C. remanei; Cb, C. briggsae. Mutations of the four groups of conserved, solvent-exposed residues (to alanines) are highlighted in different colours. (D) Coomassie stained SDS–PAGE gel showing the result of in vitro pull-down of SAS-5 by wild-type (wt) and the four mutations of SAS-6. All mutations except for mC failed to interact with SAS-5. (E) Sequence of the SAS-5 CTD. Eleven mutations are indicated as M1–M11. (F) Coomassie stained SDS–PAGE gel showing the results of in vitro pull-down of wild-type or mutants of the SAS-5 CTD by SAS-6. The four mutations that show a drastic decrease of binding to SAS-5 are indicated with arrowheads. (G) Docking the SAS-5 CTD to its binding site on the SAS-6 CCD by ClusPro 2.0 (Kozakov et al, 2010). Side chains of the residues that participate in the interaction are shown.