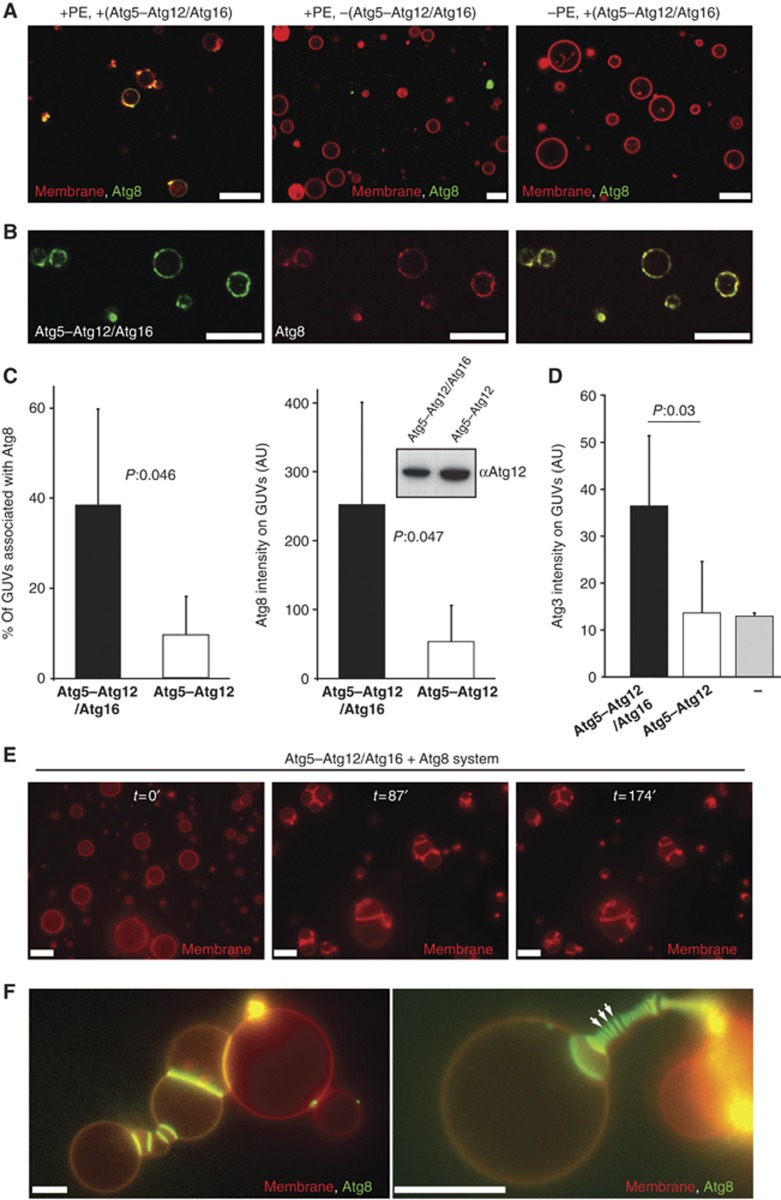
Figure 4.
Reconstitution of Atg5–Atg12/Atg16 and Atg8 on GUVs. (A) Confocal images showing eGFP–Atg8 recruitment to GUVs is dependent on the Atg5–Atg12/Atg16 complex and PE. The membrane was labelled by incorporation of rhodamine-PE. (B) Single confocal section showing colocalization of Atg5–Atg12/Atg16–eGFP and mCherry–Atg8 on GUVs. (C) Quantification of GUV-associated eGFP–Atg8 in the presence of the Atg5–Atg12/Atg16 complex and the Atg5–Atg12 conjugate. The first graph shows the percentage of GUVs associated with eGFP–Atg8 fluorescence. The second graph shows the intensity of eGFP–Atg8 on the GUVs. The quantification is based on four independent experiments. Inset: anti-Atg12 western blot using the rabbit anti-Atg12 antiserum showing the amounts of Atg5–Atg12/Atg16 complex and Atg5–Atg12 conjugate in one of the assays used for the quantification shown in (C). (D) Quantification of membrane recruitment of Atg3–mCherry by the Atg5–Atg12/Atg16 complex, the Atg5–Atg12 conjugate and a buffer control. The graph is based on two independent experiments. (E) Widefield images showing individual frames of a time series of GUVs incubated with Atg3, Atg7, Atg8 and the Atg5–Atg12/Atg16 complex (see Materials and methods for details). The membrane was labelled by incorporation of rhodamine-PE. The time is indicated in minutes. (F) Widefield images of GUVs incubated with Atg3, Atg7, eGFP–Atg8 and the Atg5–Atg12/Atg16 complex for 35 min (left) or 70 min (right). The membrane was labelled by incorporation of rhodamine-PE. Note the massive accumulation of eGFP–Atg8 at the interface between individual GUVs. The arrows point to some of these interfaces. For the quantifications, the averages and standard deviations are shown. P-values were calculated using Student’s t-test. Scale bars: 10 μm. Figure source data can be found with the Supplementary data.