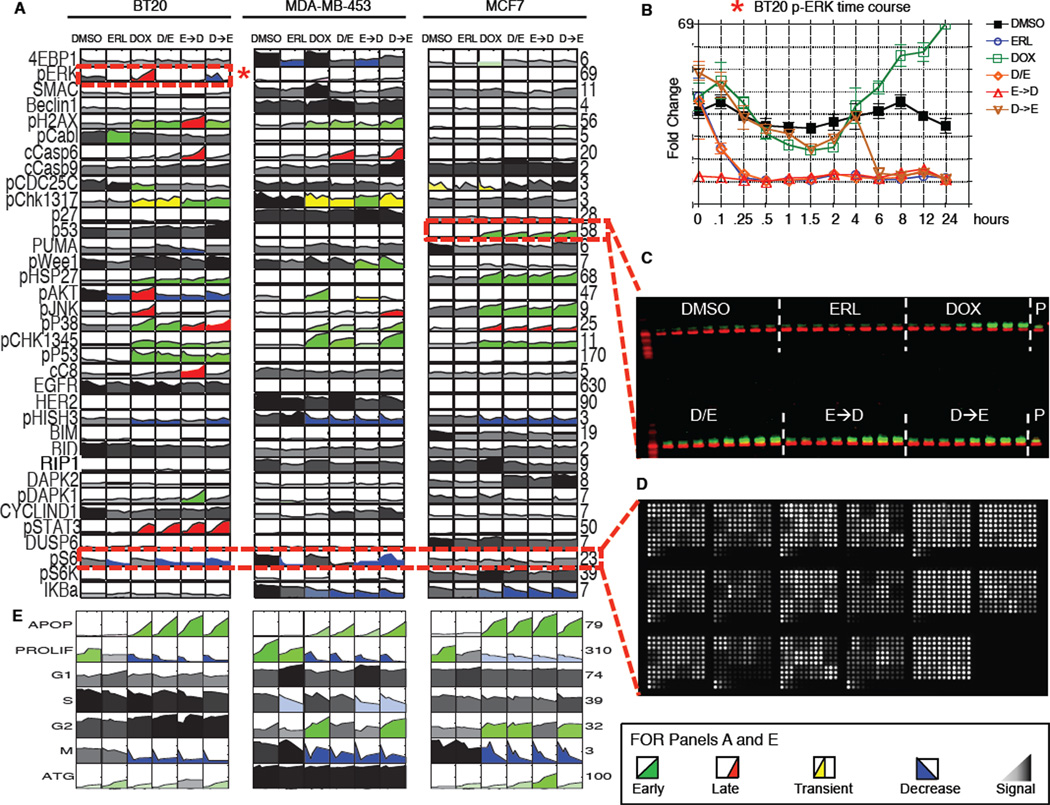
Figure 4. A systems level signal-response dataset collected using a variety of high-throughput techniques.
(A–D) (A) The complete signaling dataset for 3 different breast cancer sub-types following combined EGFR inhibition and genotoxic chemotherapy treatments as in Figure 1. Each box represents an 8- or 12-point time course of biological triplicate experiments. Time course plots are colored by response profile, with early sustained increases in signal colored green, late sustained increases colored red, and transient increases colored yellow. Decreases in signal are colored blue. Signals that are not significantly changed by treatment are shaded grey to black with darkness reflecting signal strength. Numbers to the right of each plot report fold-change across all conditions/cells. (B) Sample detailed signaling time course from panel A, highlighted by dashed box and asterisk, showing p-ERK activation in BT-20cells. Mean values ± S.D. of 3 experiments shown. (C) 48-sample Western blots analyzed using 2-color infrared detection. Each gel contained an antibody-specific positive control (P) for blot-toblot normalization. The example shown is one of three gels for total p53 in MCF7 cells (p53 in green; β-actin in red). (D) Reverse phase protein lysate microarrays were used to analyze targets of interest when array-compatible antibodies were available. The slide shown contains ~2,500 lysate spots (experimental and technical triplicates of all of our experimental samples, and control samples used for antibody calibration), probed for phospho-S6. (E) The complete cellular response dataset, colored as in A.