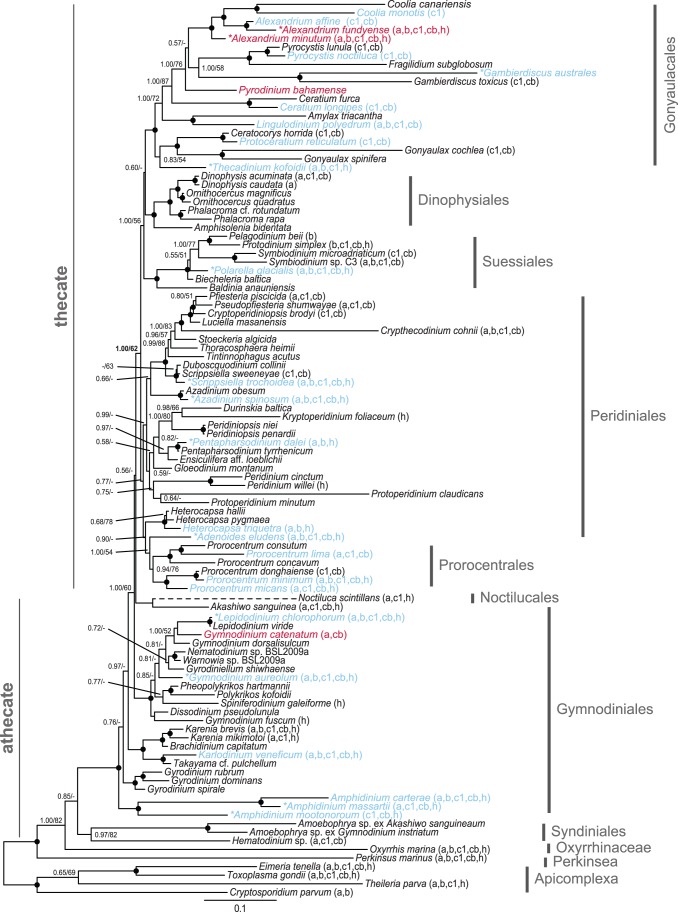
Figure 3. Phylogenetic tree of dinoflagellates inferred from rDNA, mitochondrial and nuclear protein genes.
Concatenated phylogeny, inferred from 18S+5.8S+28S+cob+cox1+actin+beta-tubulin+hsp90 (7138 characters). The tree is reconstructed with Bayesian inference (MrBayes). Numbers on the internal nodes represent posterior probability and bootstrap values (>50%) for MrBayes and RAxML (ordered; MrBayes/RAxML). Black circles indicate a posterior probability value of 1.00 and bootstrap >90%. N. scintilans is represented with a dashed branch as this taxon was excluded from the inference; alternatively its most “probable” placement was determined from a parallel Bayesian analysis. The cytochrome genes cob and cox1 for H. triquetra were excluded from the inference, a parallel phylogeny including these genes for this taxon can be seen in Figure S1. * Denotes taxa sequences generated from this study. See Table S2 for a full listing of accessions used. Red font indicates sxtA presence and blue font indicates no sxtA detection. Non-ribosomal gene presence for each taxon is represented in brackets behind each species name (a: actin, b: beta-tubulin, c1: cox1, cb: cob, h: hsp90). The phylogenetic support for the thecate/athecate split is highlighted with bold type.