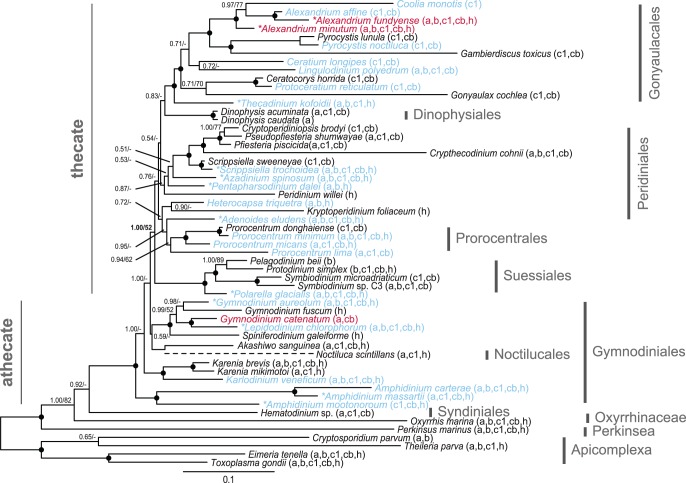
Figure 4. Phylogenetic tree of dinoflagellates inferred from rDNA, mitochondrial and nuclear protein genes (reduced phylogeny).
Concatenated phylogeny, inferred from 18S+5.8S+28S+cob+cox1+actin+beta-tubulin+hsp90 (7138 characters). This phylogeny was inferred excluding taxa with only rDNA signal; done to evaluate effects of missing characters and taxon sampling on the inference shown in Fig. 3. The tree is reconstructed with Bayesian inference (MrBayes). Numbers on the internal nodes represent posterior probability and bootstrap values (>50%) for MrBayes and RAxML (ordered; MrBayes/RAxML). Black circles indicate a posterior probability value of 1.00 and bootstrap >90%. N. scintilans is represented with a dashed branch as this taxon was excluded from the inference; alternatively its most “probable” placement was determined from a parallel Bayesian analysis. The cytochrome genes cob and cox1 for H. triquetra were excluded from the inference. * Denotes taxa sequences generated from this study. See Table S2 for a full listing of accessions used. Red font indicates sxtA presence and blue font indicates no sxtA detection. Non-ribosomal gene presence for each taxon is represented in brackets behind each species name (a: actin, b: beta-tubulin, c1: cox1, cb: cob, h: hsp90). The phylogenetic support for the thecate/athecate split is highlighted with bold type.