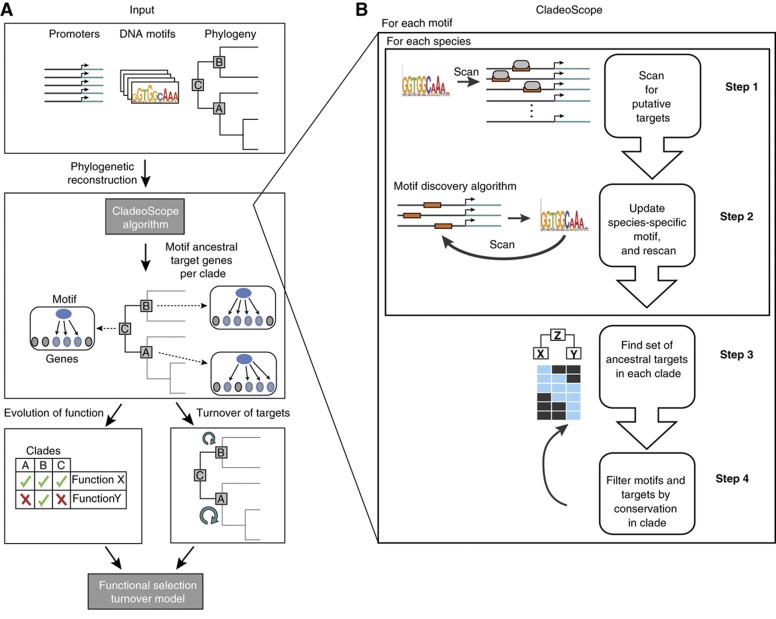
Figure 1.
The CladeoScope method. (A) Analysis overview. We use the CladeoScope algorithm that takes as input DNA motifs, promoter sequences, and the species phylogeny (top box) to reconstruct regulatory networks (i.e., ancestral motif-target genes) at each ancestral position (clade) of the phylogeny (middle box). A gene is considered as a putative target of the motif if its promoter contains an occurrence of the DNA motif (a binding site), and the ancestral motif targets are inferred by phylogenetic reconstruction. We use these networks to study both the turnover (gain and loss) of target genes associated with the motif across the phylogeny (bottom right box), and the evolution of functions associated with the motif (bottom left box) in each ancestral position of the phylogeny, where the function is determined by the functional annotation of its motif targets. We then built an evolutionary model that explains both trends. (B) The CladeoScope method. Shown is a flowchart of the input to CladeoScope (top) and its three consecutive steps: step 1—Initialization using known DNA motifs from a model organism and promoter sequences of other species. For each motif, we find putative sets of motif-containing target genes in the other genomes; step 2—Learning species-specific motifs and targets; and step 3—Network refinement, definition of detectable motifs and sets of ancestral targets per clade. step 4—Filtration of motif and target genes based on their phylogenetic conservation.