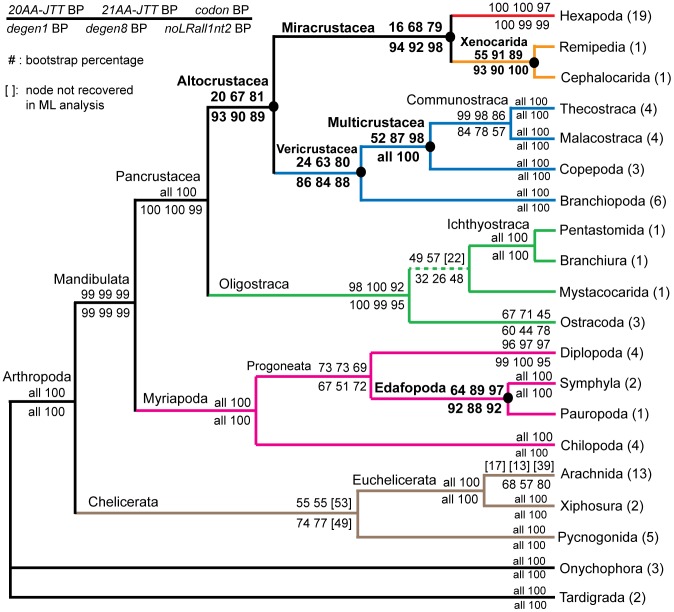
Figure 1. Deep-level arthropod relationships based on six analytical approaches.
Aligned sequences from 75 arthropods and five outgroup species for 62 nuclear protein-coding genes were analyzed under the likelihood criterion using six strategies: 20AA-JTT, a 20-amino-acid JTT model [12]; 21AA-JTT, a 21-amino-acid JTT model; codon, a codon model; degen1; degen8; noLRall1nt2. These strategies are described in the Data Set Encoding section of Materials and Methods and in [5], [6]. Numbers of species representing terminal taxa are in parentheses. Bootstrap percentages (BP) are on internal branches (20AA, 21AA, codon, degen1, degen8, and noLRall1nt2; see figure key for order). Six nodes with a major increase in their bootstrap support from 20AA JTT to 21AA JTT are identified with filled circles. A more complete listing of results can be found in Table S1.