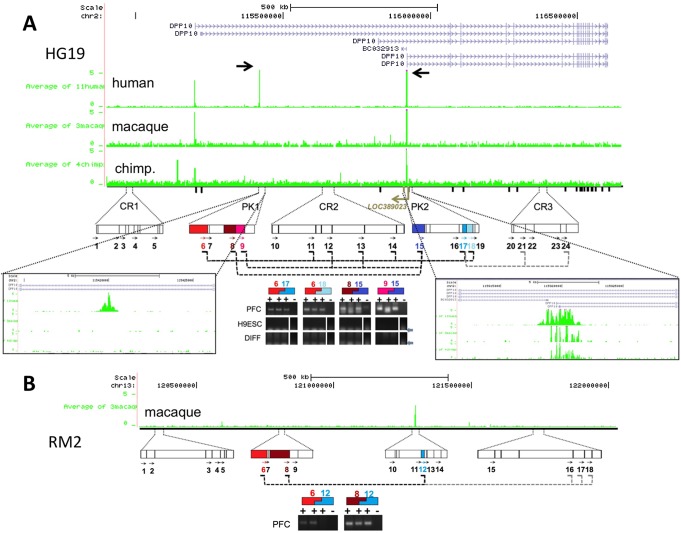
Figure 3. H3K4me3 landscapes and higher order chromatin at DPP10 (2q14.1).
(A) (Top) Genome browser tracks showing ChIP-seq H3K4me3 signal at DPP10 locus annotated to HG19 and RM2 genomes. Data expressed as normalized tag densities, averaged for 11 humans, four chimpanzees, and three macaques as indicated (see also Figure S1 for comparative annotation for each of the 18 specimens in HG19 at DPP10/2q14.1, and for the non-human primates also for the homologous loci in their respective genomes, PT2 and RM2). Human-specific peak DPP10-1 (1,455 bp) and DPP10-2 (3,808 bp) marked by arrows and shown at higher resolution in boxes, as indicated. (Bottom) Rectangles and arrows mark Hind III restriction fragments and primers from DPP10-1/2 (PK1, 2) and control regions (CR1-3) for 3C assays (human). Dotted lines connect primer pairs with sequence-verified product, indicating physical interaction of the corresponding fragments. Agarose gels for representative PCR products from 3C with (+) or without (−) DNA ligase (human primers 6,17: 282 bp; 6,18: 423 bp; 8,15: 160 bp; 9,15: 130 bp). (B) Rectangles and arrows mark Hind III restriction fragments and primers for corresponding DPP10 sequences in RM2, for macaque brain 3C. Macaque primers 6,12:298 bp, 8,12:154 bp. Notice positive interaction of PK1 with PK2 and neighboring CR2, but with not CR1 or CR3. Notice no signal in PFC 3C assays without DNA ligase and no signal in all 3C assays from H9 pluripotent (H9ESC) and differentiated (DIFF) cell cultures.