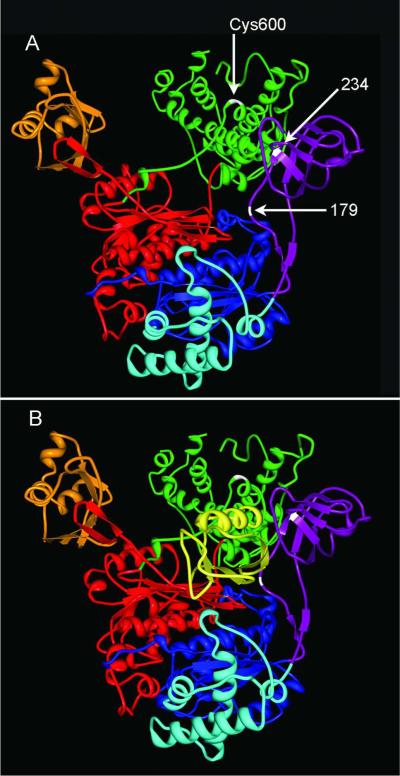
Figure 6.
Relative locations of adducted residues. Neither human nor rat E1 structures were available at the time of writing but computer modeling has supported a high degree of similarity between the quarternary structures of human E1 (P22314) and Saccharomyces cerevisiae E1 (22515).47 This yeast E1 has ~50% overall sequence homology and 88% sequence homology within the ubiquitin binding region with human E1 and rat E1. (A) The structure of yeast E1 activating enzyme was obtained from the Protein Database (PDB ID: 3CMM), and is presented as a model for canonical E1 structure. The common domains of E1s are colored as follows: the inactive adenylation domain (IAD) in blue, the active adenylation domain (AAD) in red, ubiquitin fold domain (UFD) in orange, the four helix bundle domain (4HB) in aqua, the first catalytic cysteine half domain (FCCH) in purple, and the second catalytic cysteine half domain (SCCH) in green. The S-(ethylaminothiocarbonyl) adducts on amino acid residues 179 and 234 were located within the FCCH domain of rat E1 (purple). The active site cysteine Cys600 (white) is located in the SCCH domain. (B) The decreased levels of activated E1 obtained from DEDC exposed animals suggest that covalent modification of residues within the FCCH region by DEDC may inhibit either the binding or adenylation of ubiquitin (Ub), shown in yellow, or formation of the thioester linkage of Ub to Cys600.