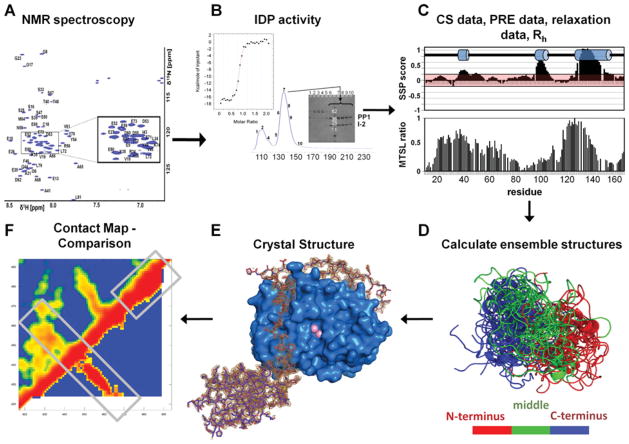
Figure 1. Typical IDP analysis workflow.
(A) NMR spectroscopy can be readily used to identify IDPs. Two-dimensional 1H,15N HSQC (heteronuclear single-quantum coherence) spectrum of IDPs have a limited peak dispersion in the 1HN dimension due to a lack of hydrogen-bonding network typical in secondary-structural elements of well-folded proteins. (B) Necessary experiments to verify protein function. For this system, IDP function is tested by IDP–PP1 complex formation, either by co-elution from a size-exclusion chromatography column or by isothermal titration calorimetry. (C) Measurement and evaluation of chemical shift (CS), PRE, auto-correlated short-timescale relaxation NMR data as well as global parameters, based on dynamic light scattering or SAXS. MTSL, S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate spin label; Rh, hydrodynamic radius; SSP, secondary-structure propensity. (D) Calculation of the IDP ensemble structure. (E) Determination of the complex structure between the IDP and the targeting protein (spinophilin–PP1 structure is shown; PDB code 3EGG) using X-ray crystallography or NMR spectroscopy. (F) Comparison of unbound and bound IDP conformations via a distance correlation map, to evaluate the mode of interaction, as well as differential and common structural features.