Abstract
The stability of pathogen-specific DNA or RNA amplification targets in clinical samples following short-term storage at room temperature, 4°C, and −80°C was assessed by real-time PCR. In purified nucleic acid extracts, both DNA and RNA targets were stable for up to 30 days, irrespective of storage temperature. In unextracted samples, temperature-dependent loss of targets (P < 0.05) was observed in serum and cerebrospinal fluid specimens, while no changes were observed for EDTA blood samples.
TEXT
Nucleic acid amplification tests (NAATs) to detect infectious pathogens are now widely used in clinical microbiology laboratories. Although these assays provide high sensitivity, high specificity, and rapid turnaround time, special laboratory practice and procedures are required to avoid false-positive and false-negative results (1, 2). Optimal application of NAATs depends on a number of preanalytical factors, such as specimen collection, transport, and storage conditions (3, 4). These factors impact the stability of pathogen-associated nucleic acids (NA), which must be protected from physical, chemical, or enzymatic degradation during specimen transport and handling. The most important threats to the stability of NA targets in clinical samples are nucleases, such as DNases and RNases, which may be released from lysed human cells and microorganisms (5). In real-life clinical situations, questions arise over whether clinical samples or NA extracts, left at room temperature (RT) for a few days or stored in the refrigerator for a week, are still valid for molecular testing. Although there are published guidelines for the collection, transport, and storage of specimens for molecular tests, described by CLSI (4), they are based mostly on opinions and limited experimental data. Published data on the stability of pathogen-specific amplification targets are also limited to individual pathogens and/or specimen types (3, 4, 6–10, 12). Therefore, in this study, we undertook a broader study to compare the stability of extracted and unextracted clinical samples over time and at different temperatures.
To determine the short-term storage stability of purified DNA and RNA, NA was extracted from known positive specimens containing the following: Neisseria meningitidis in EDTA blood, cytomegalovirus (CMV) and Epstein-Barr virus (EBV) in serum, and influenza A virus in nasopharyngeal washes (NPW). All samples were extracted using the QIAsymphony virus/bacteria kit in an automated DNA extraction platform QIAsymphony SP (Qiagen) except for EDTA blood, which was extracted manually using the QIAamp DNA blood minikit (Qiagen). Purified eluates were stored in aliquots under sterile conditions at room temperature (RT) (22 to 25°C), 4°C, and −80°C for 0 to 30 days.
For unextracted samples, the target pathogen and the specimen types were chosen so that they (i) represent the most common specimen type for the respective pathogen and (ii) had not previously been studied. EDTA blood, serum, cerebrospinal fluid (CSF), and NPW samples were spiked with an N. meningitidis B strain (ATCC 13090), a CMV patient isolate, a human enterovirus 70 strain (ATCC VR-836), and an influenza A virus patient isolate, respectively, within 4 h of collection (see Methods in the supplemental material). NA from spiked samples was extracted in the same way following storage at RT, 4°C, and −80°C for 0 to 16 days. Extracted nucleic acids were analyzed either by in-house (see methods and Table S1 in the supplemental material) or commercial, probe-based, real-time PCR assays. Human enterovirus RNA was detected using a commercial eQ-PCR EnV detection kit (Trimgen Corporation). For kinetic analysis, threshold cycle (CT) values for each pathogen were plotted against the number of days the samples were stored at different temperatures. The half-lives of pathogen-associated nucleic acid targets were obtained from the linear equation (see Methods in the supplemental material). For average analysis, fold changes in the relative quantity of amplification targets were calculated based on CT values using the equation FC = 2−dCT, where dCT = (CT,day x − CT,day 0) (11). The percent loss of targets was calculated from fold changes from the quantity of targets at day 0. Statistical significance was calculated by paired and 2-tailed Student's t test, and a P value of <0.05 was considered to be statistically significant. Statistical differences between multiple experimental groups were calculated by analysis of variance (ANOVA) followed by a posttest using Bonferroni correction at a 95% confidence interval.
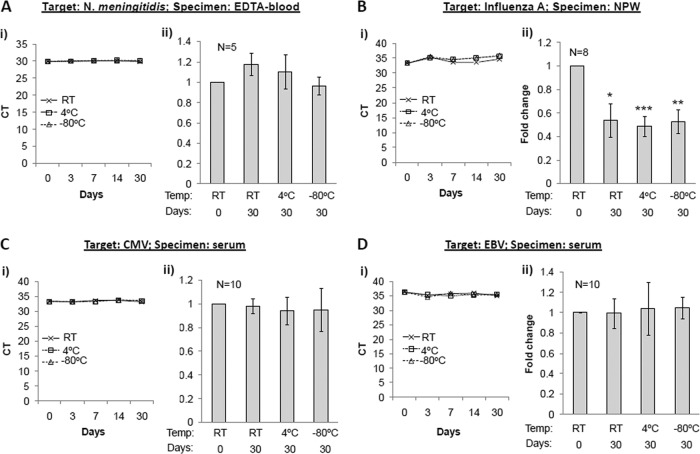
There was no loss of extracted N. meningitidis, CMV, and EBV DNA derived from EDTA blood and serum samples after storage at RT, 4°C, and −80°C for 0, 3, 7, 14, and 30 days (Fig. 1Ai, Ci, Di). To account for sample-to-sample variation, we performed further tests on additional samples at days 0 and 30 of storage at different temperature and found no significant differences in the relative quantity of specific amplification targets (Fig. 1Aii, Cii, Dii). In general, DNA, when purified and stored in a preferred buffer such as Tris-EDTA (pH 7.2), is considered to be stable for at least 1 year at 2 to 8°C and for longer terms at −70°C or lower (3). However, it is important to note that the DNA purity could be dependent on the specimen type and the extraction methods, which could subsequently influence the stability of purified DNA. Our findings suggest that DNA extracted from EDTA blood or serum samples, using an automated and magnetic bead-based extraction method, may be safely stored at RT or lower without any loss for up to 1 month. Unlike the case with DNA, it is recommended that purified RNA be stored at −70°C or lower and at a slightly alkaline pH (7.1 to 7.5) (3). In our lab setting, we observed a small (∼1 CT; P < 0.05) loss of influenza A RNA, which was not temperature dependent (between RT to −80°C), in purified NPW extracts following storage for a period of 1 month (Fig. 1B). These results indicate that temporary storage of extracted nucleic acids from clinical samples at RT or 4°C is unlikely to affect NAAT results.
Fig 1.
Storage stability of DNA and RNA in postextracted clinical samples. Nucleic acid extracts from clinical samples known positive for N. meningitidis (A), influenza A virus (B), CMV (C), and EBV (D) were assessed for respective targets by TaqMan PCR at day 0 and following storage at RT, 4°C, and −80°C for different time intervals as indicated. (Ai to Di) Kinetic data based on CT values plotted against the number of days stored at different temperature. (Aii to Dii) Average of fold changes calculated from CT values, with respect to CT values at day 0. Baseline CT values in different samples ranged from 29.9 to 38.5 for N. meningitidis, 28.4 to 39.5 for CMV, 28.2 to 38.5 for EBV, and 21.3 to 34.8 for influenza A virus. Intraassay variation between duplicate runs was <1 CT. Error bars = standard errors of the means (SEM); statistical significance was calculated by pairwise Student's t test (2-tailed); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
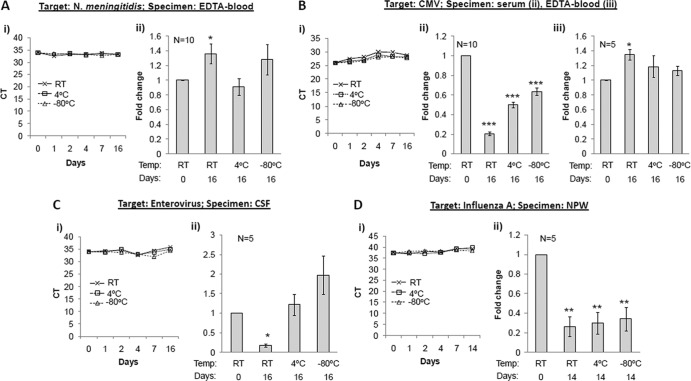
Unlike extracted samples, where purified DNA and RNA molecules are maintained in a defined buffer, crude clinical samples represent a complex biological environment, where an interplay of degradative proteases and nucleases may determine the fate of pathogen-associated amplification targets. Previously published observations on the stability of microbial DNA in clinical samples include the following: (i) Trichomonas vaginalis DNA was stable for 1 day at RT and 3 days at 4°C in urine samples (6), (ii) spiked Treponema pallidum DNA in CSF was detectable after 96 h of storage at RT and 4°C (12), (iii) HSV DNA in positive clinical specimens remained stable for at least 16 months at −20°C (7), and (iv) HBV DNA in positive plasma samples was stable at 25°C and at 5°C for at least 25 days (8), whereas in serum it was stable only for 5 days at 4°C and −70°C and less stable at RT (10). Our results on the stability of spiked N. meningitidis DNA in EDTA blood and CMV DNA in serum and EDTA blood samples correlate well and complement these observations. Whereas both N. meningitidis and CMV DNA were highly stable in EDTA blood for 2 weeks at any storage temperature used, significant loss (P < 0.05 by ANOVA and posttest) of CMV DNA was observed in serum samples in a temperature-dependent manner (Fig. 2A and B). The half-life of CMV DNA based on PCR assays was <1, 2, and 3 days when stored at RT, 4°C, and −80°C, respectively (Table 1).
Fig 2.
Storage stability of DNA and RNA in preextracted clinical samples. Clinical samples were spiked with N. meningitidis (A), CMV (B), enterovirus (C), and influenza A virus (D). Nucleic acid extraction and TaqMan PCR for the respective targets were performed on aliquots of samples at day 0 and following storage at RT, 4°C, and −80°C for different time intervals as indicated. (Ai to Di) Kinetic data based on CT values plotted against the number of days stored at different temperatures. (Aii to Dii and Biii) Average of fold changes calculated from CT values, with respect to CT values at day 0. Baseline CT values in different spiked samples ranged from 31.9 to 34.0 for N. meningitidis, 29.6 to 31.7 for CMV, 32.7 to 34.1 for enterovirus, and 33.9 to 37.1 for influenza A. Intraassay variation between duplicate runs was <1 CT. Error bars = SEM; statistical significance compared to data at day 0 calculated by pairwise Student's t test (2-tailed); *, P < 0.05; ***, P < 0.001. Multiple group comparisons for CMV DNA levels in serum samples were made by ANOVA and posttest (95% confidence interval), indicating a statistically significant (P < 0.05) difference between different groups.
TABLE 1.
Half-life and percentage loss of pathogen-specific amplification targets in different types of clinical specimens
| Pathogen | Specimen type | No. of days stored | Half-life (no. of days) |
% loss of target after 2 weeks |
||||
|---|---|---|---|---|---|---|---|---|
| RT | 4°C | −80°C | RT | 4°C | −80°C | |||
| N. meningitidis | EDTA blood | 16 | No loss | No loss | No loss | |||
| CMV | Serum | 16 | <1 | 2 | 3 | 79.7 ± 5 | 50 ± 10 | 36.9 ± 12 |
| EDTA blood | 16 | No loss | No loss | No loss | ||||
| Human enterovirus | CSF | 16 | 9 | 82.1 ± 9 | No loss | No loss | ||
| Influenza A | Nasopharyngeal aspirates | 14 | 6 | 6 | 10 | 74 ± 23 | 70 ± 25 | 66 ± 27 |
Although a loss of RNA targets in crude samples is expected during storage, unless frozen, we noted no dramatic, logarithmic decrease in human enterovirus and influenza A RNA levels in CSF and NPW samples, respectively, after 2 weeks. A small but statistically significant reduction of enterovirus RNA (65%; P < 0.05) at RT and influenza RNA (>50%; P < 0.001) at all temperatures was observed (Fig. 2C and D). The half-life of enterovirus RNA in CSF samples at RT was 9 days (Table 1), which correlates well with that of HIV-1 RNA (7 days) in plasma samples, as previously reported (8). Similarly, at 4°C, enterovirus RNA maintained its integrity for approximately 2 weeks in this study, as seen in a previous HIV-1 study (8). The results of influenza A RNA in NPW samples were somewhat surprising, as no temperature-dependent effects were detected. One limitation of this study is that we did not analyze RNA stability in EDTA blood samples. However, a previous study found HCV RNA stable in EDTA blood for up to 96 h (9), similar to what we observed for DNA targets.
In conclusion, we observed minimal loss of pathogen NAAT targets for both extracted and unextracted clinical samples within a few days of specimen collection. None of the NAAT targets became undetectable during the study period, which was limited to 1 month and 2 weeks, for pure extracts and crude samples, respectively, to represent most common scenarios in a diagnostic microbiology laboratory. The temperature-dependent loss of targets in serum and CSF samples indicates possible enzymatic degradation. In contrast, EDTA blood samples were found to be the most protective medium for nucleic acid stability. This may be because EDTA sequesters metal ions required for enzymatic activity and thus inhibits nucleases and proteases. Based on these observations, we conclude that, if a processing delay is unavoidable, EDTA blood or plasma samples are preferable to serum samples for real-time PCR detection of bacterial or viral pathogens.
Supplementary Material
Footnotes
Published ahead of print 10 October 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Belkum A. 2003. Molecular diagnostics in medical microbiology: yesterday, today and tomorrow. Curr. Opin. Pharmacol. 3:497–501 [DOI] [PubMed] [Google Scholar]
- 2. Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 23:550–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2005. Collection, transport, preparation, and storage of specimens for molecular methods; approved guideline. MM13-A CLSI, Wayne, PA [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2006. Molecular diagnostic methods for infectious diseases; approved guideline—2nd edition. MM3-A2 CLSI, Wayne, PA [Google Scholar]
- 5. Fersht AR. 1977. Enzyme structure and mechanism, 2nd edition W.H. Freeman, Reading, PA [Google Scholar]
- 6. Ingersoll J, et al. 2008. Stability of Trichomonas vaginalis DNA in urine specimens. J. Clin. Microbiol. 46:1628–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jerome KR, Huang M, Wald A, Selke S, Corey L. 2002. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J. Clin. Microbiol. 40:2609–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jose M, Gajardo R, Jorquera JI. 2005. Stability of HCV, HIV-1 and HBV nucleic acids in plasma samples under long-term storage. Biologicals 33:9e16. [DOI] [PubMed] [Google Scholar]
- 9. Kessler HH, et al. 2001. Effects of storage and type of blood collection tubes on hepatitis C virus level in whole blood samples. J. Clin. Microbiol. 39:1788–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krajden M, et al. 1998. Assessment of hepatitis B virus DNA stability in serum by the chiron quantiplex branched-DNA assay. J. Clin. Microbiol. 36:382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Livac KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 12. Villanueva AV, Podzorski RP, Reyes MP. 1998. Effects of various handling and storage conditions on stability of Treponema pallidum DNA in cerebrospinal fluid. J. Clin. Microbiol. 36:2117–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.