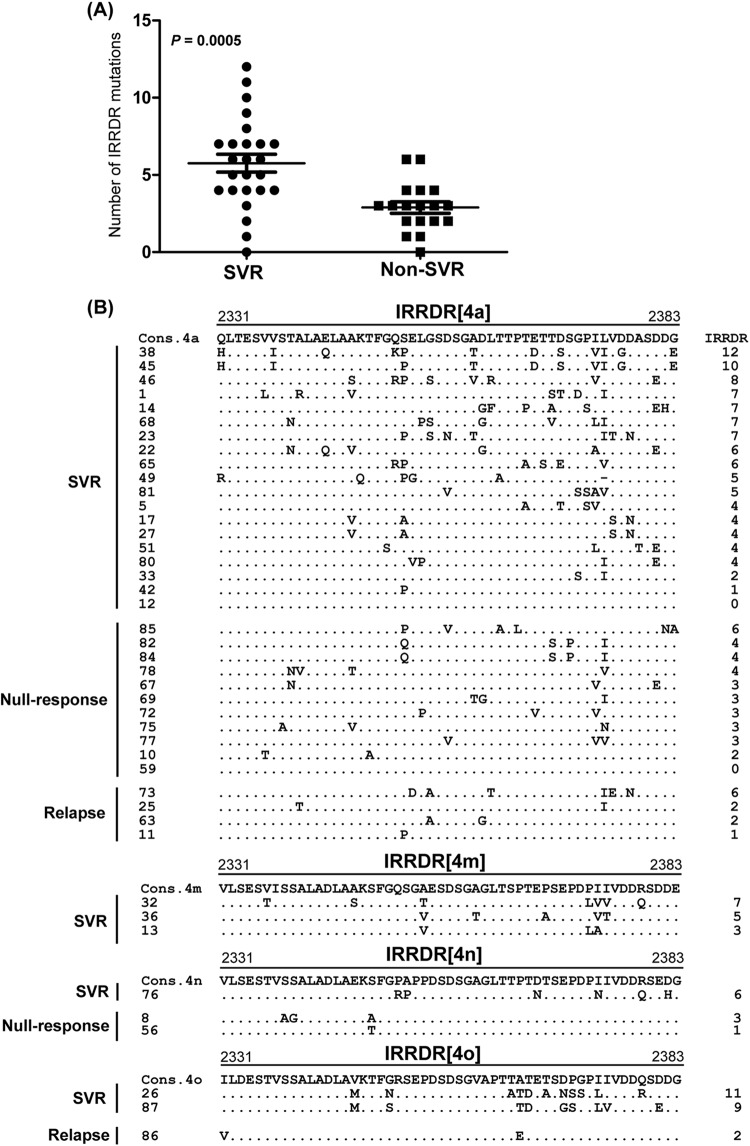
Fig 2.
Correlation between IRRDR[HCV-4] sequence variations and treatment outcome. (A) Average number of amino acid mutations in IRRDR[HCV-4] obtained from SVR and non-SVR patients. (B) Alignment of IRRDR[HCV-4] sequences obtained from SVR and non-SVR patients with HCV-4a, -4m, -4n, and -4o. The consensus sequence (Cons) of each subtype is shown on the top. The numbers along the sequence indicate the amino acid positions. Dots indicate residues identical to those of the Cons sequence. The numbers of the mutations in each IRRDR (4a, 4m, 4n, or 4o) are shown on the right.