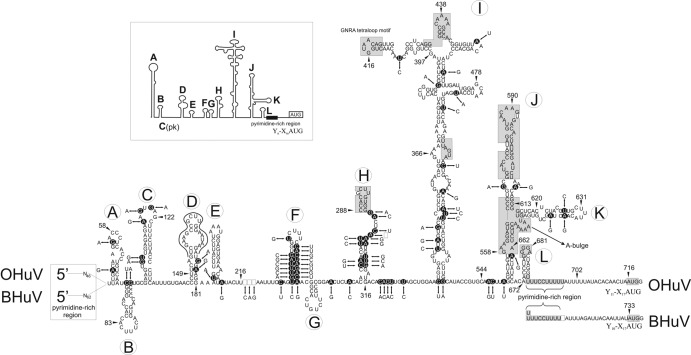
Fig 2.
Predicted RNA secondary structure of the OHuV-1 (HM153767) 5′ UTR, including the IRES, as determined using the Mfold program. The RNA secondary structure of OHuV-1 is supplemented with the BHuV-1 (JQ941880) 5′ UTR nucleotide sequence for comparison. The positions of nucleotide differences are indicated by double arrows between OHuV-1 (dark circles) and BHuV-1; however, the pyrimidine-rich region at the extreme 5′ end and the Yn-Xm-AUG motif, which corresponds to the lengths of the pyrimidine tract/spacer sequence/AUG initiation codon at the 3′ end of the 5′ UTR, are indicated for both viruses. Nucleotide deletions/insertions are indicated by empty squares. The complete structure of the 5′ UTR, including the domains from A to L and the type II IRES, has been annotated as previously proposed for human parechoviruses (inset) (9). The central five IRES domains are labeled from H to L to maintain the continuity of the current nomenclature. The positions of conserved type II IRES motifs; the nucleotides with high identity to human parechoviruses in IRES domains H, I, J, and L; the pyrimidine-rich region at the 3′ end; and the predicted polyprotein AUG start codon are indicated by shaded boxes. In domain D, the continuous black line shows the positions of the 22 nucleotides that are identical in hungaroviruses and Ljungan virus 4 (EU854568).