Abstract
Methylation of the high-risk human papillomavirus type 16 (HPV16) upstream regulatory region (URR) has been described, but whether methylation is present among low-risk HPVs is unknown. The methylation status of the HPV6 URR was analyzed in papillomas from the upper aerodigestive tract of six adult patients. All CpGs in the URR were unmethylated, from both basal/intermediate and superficial cells, suggesting that methylation is not involved in the regulation of transcription from the HPV6 URR, regardless of epithelial differentiation.
TEXT
Papillomaviruses are small icosahedral viruses with an ∼8-kb double-stranded DNA circular genome, and more than 150 human papillomavirus (HPV) types have been characterized to date (1). These viruses are epitheliotropic and produce hyperproliferation of squamous cells, generally referred to as papillomas. High-risk HPV types are associated with cancer, especially of the uterine cervix, while low-risk types produce benign warts (29).
Methylation of viral genomes of high-risk HPVs has been identified as an important feature in cervical neoplasms (5, 16, 28). It has been speculated that methylation regulates the viral gene expression in multicopy HPVs (3) as well as the binding of the regulatory protein E2 to the E2 binding sites in the URR (11). Differential methylation has been observed in the URR of HPV type 16 (HPV16) during epithelial differentiation, where the p97 promoter region was highly methylated in superficial cells, while the basal and intermediate layers were methylated mainly in the enhancer region of the URR (28). Furthermore, methylation of the HPV16 URR inhibits transcriptional activation by the HPV E2 protein (11), but whether methylation is present in low-risk HPVs is unknown. Therefore, our aim was to evaluate the methylation status of the viral genomes in the low-risk HPV6. We analyzed HPV6 in papillomas of the upper aerodigestive tract of adult patients. These benign lesions often recur and can have significant morbidity and, rarely, mortality as a consequence of airway obstruction (6, 13).
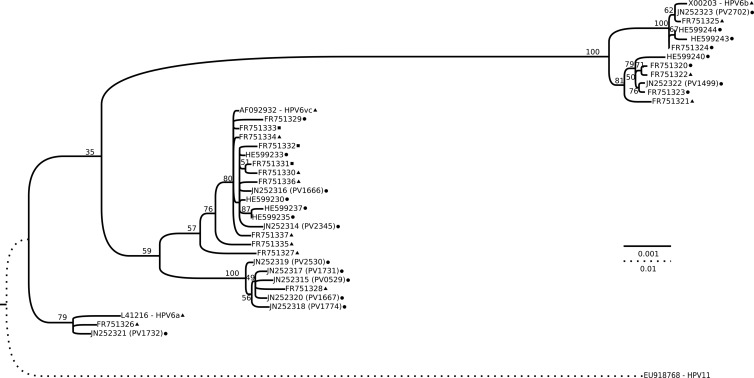
In order to design primers suitable for methylation analyses, we performed a complete genome characterization of our HPV6 isolates, originally identified using PCR with modified GP5+/6+ (MGP) primers (23) and a Luminex system (19). The HPV6 isolates were from patients ages 24 to 68 years who attended otorhinolaryngology clinics in Sweden. We selected one pharyngeal and nine laryngeal HPV6-positive biopsy specimens (Table 1). In order to amplify the complete HPV6 genomes, automated DNA extraction (MagNA Pure LC total nucleic acid isolation; Roche) and long-range PCR (Expand Long Template PCR system; Roche) were performed (Table 2) (27). By using long PCR products as the templates, the complete genomic sequences were obtained by using 15 sequencing primers (Table 2). In order to establish the phylogenetic relationships, a multiple sequence alignment was made using the MUSCLE version 3.8 plug-in (7) from the UGENE version 1.10 package (15). From the alignment, a maximum likelihood phylogenetic tree (Fig. 1) was constructed using the RAxML software (version 7.3.0) (24) under the raxmlGUI program (version 1.1) (21). The general time-reversible (GTR) model of nucleotide substitution under the gamma model of rate heterogeneity was selected for 20 inferences with 800 bootstraps. HPV type 11 (accession number EU918768) was used to root the tree, and the FigTree software (version 1.3.1) (18) was used to create a graphical representation of the tree.
TABLE 1.
Nucleotides and amino acids of 10 HPV type 6 isolates in laryngeal papillomas versus the closest reference sequence
| Isolate no. | Patient characteristics |
URR metha | Closest HPV (accession no.) | Size (bp) | Accession no. | No. (%) of different nucleotides | No. of indicated mutation type foundb |
Amino acid conservationc |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (yrs) | Sex | Smoker? | Site | Missense | Silent | Ins | Del | Ts | Tv | NC | WC | SC | ||||||
| PV2702 | 67 | F | Never | Larynx | No | HPV6b (X00203) | 7996 | JN252323 | 2 (0.03) | 0 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| PV1499 | 43 | M | Never | Larynx | No | HPV6b (X00203) | 7996 | JN252322 | 18 (0.23) | 0 | 19 | 1 | 0 | 8 | 10 | 0 | 0 | 0 |
| PV1666 | 68 | M | Never | Larynx | NA | HPV6vc (AF09293) | 8031 | JN252316 | 4 (0.05) | 2 | 3 | 1 | 0 | 2 | 2 | 1 | 1 | 0 |
| PV2345 | 40 | M | Never | Larynx | No | HPV6vc (AF09293) | 8032 | JN252314 | 6 (0.07) | 3 | 7 | 1 | 0 | 3 | 3 | 1 | 1 | 1 |
| PV0529 | 58 | M | Quit | Pharynx | No | HPV6vc (AF09293) | 8031 | JN252315 | 30 (0.37) | 12 | 19 | 1 | 0 | 14 | 16 | 2 | 4 | 6 |
| PV1667 | 39 | M | Never | Larynx | NA | HPV6vc (AF09293) | 8029 | JN252320 | 29 (0.36) | 11 | 20 | 1 | 1 | 14 | 15 | 2 | 4 | 5 |
| PV1731 | 40 | M | Never | Larynx | NA | HPV6vc (AF09293) | 8031 | JN252317 | 29 (0.36) | 11 | 19 | 1 | 0 | 14 | 15 | 2 | 4 | 5 |
| PV1774 | 56 | M | Never | Larynx | No | HPV6vc (AF09293) | 8031 | JN252318 | 31 (0.39) | 12 | 20 | 1 | 0 | 16 | 15 | 2 | 4 | 6 |
| PV2530 | 40 | M | Never | Larynx | No | HPV6vc (AF09293) | 8031 | JN252319 | 28 (0.35) | 11 | 18 | 1 | 0 | 15 | 13 | 2 | 3 | 6 |
| PV1732 | 24 | M | Quit | Larynx | NA | HPV6a (L41216) | 8010 | JN252321 | 11 (0.14) | 4 | 7 | 0 | 0 | 8 | 3 | 3 | 0 | 1 |
Methylation status in the URR. NA, not analyzed.
Ins, insertion; Del, deletion; Ts, transition; Tv, transversion.
NC, nonconserved amino acid change; WC, weakly conserved amino acid change; SC, strongly conserved amino acid change.
TABLE 2.
Primers and PCR settings
| Primer name | Sequence (5′–3′) | Use | Cycling conditions | Vol (μl) | Mg2+ concn (mM) | dNTP concn (mM) | Fwd primer concn (μM) | Rev primer concn (μM) | BSA concn (%) | Enzyme units | Template vol (μl) | Enzyme systema |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV 6 6125F | GGGTGTAAGTGGACATCCTTTCCTA | Long-range PCR | 94°C 2 min; 20 cycles of 94°C 10 s, 61°C 30 s, 68°C 6 min; 24 cycles of 94°C 15 min, 59°C 30 min, 68°C 6 min 10 s; 68°C 10 min | 50 | Buffer 2 | 0.5 | 0.3 | 0.3 | 0.00 | 1.78 | 5 | Expand L-T |
| HPV 6 6124R | ACACCTAATGGCTGTCCCCTG | |||||||||||
| HPV 6 116F | ATGCCTCCACGTCTGCAACGACC | Long-range PCR (alternative) | 94°C 2 min; 10 cycles of 94°C 10 s, 68°C − 0.5°C/cycle (to 63°C) 30 s; 68°C 6 min; 10 cycles of 94°C 10 s, 61°C 30 s, 68°C 6 min; 24 cycles of 94°C 15 s, 59°C 30 s, 68°C 6 min 10 s; 68°C 10 min | 50 | Buffer 2 | 0.5 | 0.3 | 0.3 | 0.00 | 1.78 | 5 | Expand L-T |
| HPV 6 115R | TTGCACTTTCCATAATGCCTCGTTTGCT | |||||||||||
| Met URR 1F_b | TWRTTATATTTTGTGATTTAGTGGTTGTTGTA | URR methylation study set 1 (5′ segment) | 94°C 5 min; 10 cycles of 94°C 15 s, 60°C 20 s − 1°C/cycle, 72°C 25 s; 45 cycles of 94°C 15 s, 60°C 20 s, 72°C 25 s; 72°C 5 min | 20 | 3.5 | 0.2 | 0.3 | 0.9 | 0.20 | 0.5 | 2 | AmpliTaq Gold |
| Met URR 1R_b | AACACATTATAACAAATTAATAMAAAATATATACYAAAAACA | |||||||||||
| Met URR 2F | GGTTGTTTTTRGTATATATTTTKTATTAATTTGTTAT | URR methylation study set 2 (central segment) | 94°C 5 min; 10 cycles of 94°C 15 s, 60°C 20 s − 1°C/cycle, 72°C 25 s; 45 cycles of 94°C 15 s, 55°C 20s, 72°C 25 s; 72°C 5 min | 20 | 3.5 | 0.2 | 0.3 | 0.3 | 0.20 | 0.5 | 2 | AmpliTaq Gold |
| Met URR 2R | AATTAACTACAATACATAAAAATATAACAC | |||||||||||
| Met URR 3F_b | GTTTGGTATATAATAATATAAAAATGAGTAATTTAAGGTTATAT | URR methylation study set 3 (3′ segment) | 94°C 5 min; 10 cycles of 94°C 15 s, 60°C 20 s − 1°C/cycle, 72°C 25 s; 45 cycles of 94°C 15 s, 60°C 20 s, 72°C 25 s; 72°C 5 min | 20 | 3.5 | 0.2 | 0.3 | 0.3 | 0.20 | 0.5 | 2 | AmpliTaq Gold |
| Met URR 3R_b | TTACAACATATACATAAATAAATTAAACATCTTACACAAC | |||||||||||
| HPV16 Met 7.2k F | ATTATTGTGTTATGTAATATAAATAAATTT | Methylation positive control in CaSki cells | 94°C 5 min; 10 cycles of 94°C 15 s, 60°C 20 s − 1°C/cycle, 72°C 25 s; 45 cycles of 94°C 15 s, 48°C 20 s, 72°C 25 s; 72°C 5 min | 30 | 3.5 | 0.2 | 0.3 | 0.3 | 0.20 | 0.75 | 3 | AmpliTaq Gold |
| HPV16 Met 7.5k R | AATCAAAAAAACAAAAATTTAACAC | |||||||||||
| E6 90F | CARACGAGGCATTATGGAAAG | Sequencing primer for HPV6 | Sequenced at MWG (Germany) with ABI 3730XL equipment | |||||||||
| E6 224R | GCAGTGGTCAGTGCATTCYTG | |||||||||||
| E7 688F | AAATASTGACCTGTTGCTGTG | |||||||||||
| E1 1898R | ATTGACTGTCAGCCAACCYAT | |||||||||||
| E2 2878R | CTYAGGCCCATTTGTTTTGCT | |||||||||||
| E1 1681F | AGTAAATAAAAGTAGATGTACCGTKGC | |||||||||||
| E2 HPV-6 3819R | GCATTGACATAAACCCCAGTTT | |||||||||||
| E1 2598F | TTCCAAATCCATTCCCYTTTG | |||||||||||
| E5b 4558R | TAATATTTGATCTGCAATRGT | |||||||||||
| E4 3486F | CACATTAGATCCGTGGACAGT | |||||||||||
| HPV 6 4826R | CAGAGGATGTAATTGTAAACCCACC | |||||||||||
| E5b 4176F | AATTTAATGATGGTGATACMTG | |||||||||||
| HPV 6 4680F | CTATGGCTCGTCCTCCTGTGG | |||||||||||
| HPV 6 6328 F | ACTGCCCGCCCTTAGAACTTATT | |||||||||||
| HPV 6 7714R | CAAAATATATAGGATTAACAGGCAAC |
The system used was either the Expand Long–Template (Expand L-T) PCR system or the AmpliTaq Gold system (both obtained from Roche), unless sequencing was performed at MWG with ABI 3730XL equipment.
Fig 1.
Phylogenetic tree based on the complete genomes of all unique HPV6 variants available in GenBank. All accession numbers are indicated in the leaves of the tree, and for our 10 new HPV6 genomes the isolate name is given in parentheses. The isolation source is indicated, as follows: circle, laryngeal papilloma; triangle, condyloma acuminata; square, both laryngeal papilloma and condyloma acuminata (for genomes deposited in GenBank with identical sequences but different isolation sites). The tree was obtained by the maximum likelihood approach using RAxML software and rooted with HPV11. Bootstrap (800) support values are indicated in each branch as percentages. The bars indicate the number of substitutions per site for the two different scales used.
The characteristics of the different genomes are summarized in Table 1. The average nucleotide identity by pairwise comparison between our isolates was 99.1% (range, 98.1% to 99.9%). We also inferred the phylogenetic relationships of HPV6 by using the complete genomes of our 10 isolates and the 28 unique HPV6 sequences available in GenBank. Of our HPV6 isolates, seven clustered with HPV6vc (in two subclusters), two clustered together with HPV6b, and one clustered with HPV6a (Fig. 1). When taking into account another 25 HPV6 isolates deposited in GenBank with sequences identical to those present in the tree, the HPV6vc-related variants were the most prevalent isolates both in genital (65% [11/17]) and oral (58% [21/36]) lesions. This common occurrence of HPV6vc-related variants is in agreement with a previous study (12). It is noteworthy that more than 30 years after the HPV6b prototype was isolated (4, 20), we have now identified an HPV6b variant (PV2702) with the 94-bp insert and only two point mutations, supporting the supposition that the original 94-bp deletion was an artifact (9).
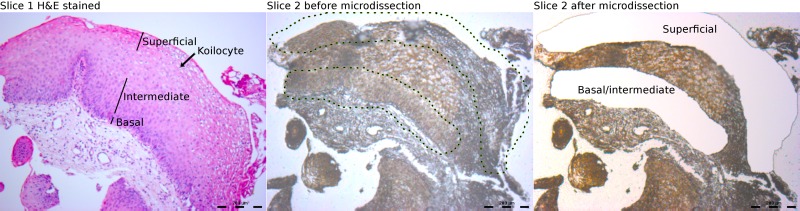
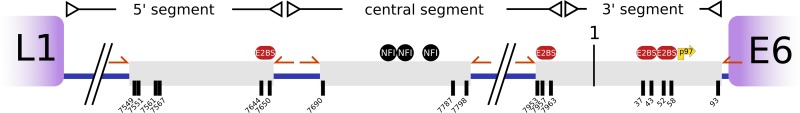
As the URR is critical for replication and transcription, we focused our methylation exploratory study on this region of the genome. We analyzed the basal/intermediate cells separately from the superficial cells of six HPV6-positive samples (Table 1). The biopsy specimens were trimmed, serially dehydrated, and paraffin embedded. Due to the small size of the biopsy specimens, it was not possible to stain the slices when they were mounted on special membranes for laser capture microdissection, as large portions were lost during ethanol washes. To circumvent this problem, eight consecutive slices were cut (5 μm thick), of which only the first and last were mounted on glass slides and stained with hematoxylin and eosin. The remaining sections were mounted on polyethylene membranes on steel frames (PET; Leica) without staining. The first and last slides were used as references to identify basal and superficial regions on the PET slides. Microdissection was performed with an LMD6000 microdissection system (Leica) following the manufacturer's instructions (Fig. 2). DNA extraction from the basal/intermediate and superficial fractions was performed using the RecoverAll total nucleic acid isolation kit (Ambion). Initially, the slices were suspended in 0.5 ml of xylene and then mixed with 1 ml of ethanol to facilitate recovery after centrifugation. The purification was completed following the manufacturer's instructions. Afterwards, the purified DNA was bisulfite treated using the EZ DNA methylation kit (Zymo Research). CaSki cervical cancer cells obtained from the American Type Culture Collection were treated in parallel and employed as a positive control of methylation, using primers published elsewhere (Table 2) (5). Three segments of the URR of HPV6 were amplified using primers insensitive to methylation (without CpGs), which included all the CpGs and E2 binding sites present in that region of the genome (Fig. 3; Table 2). Ten clones of each amplicon, from both basal/intermediate and superficial cells, were sequenced to identify possible methylated sites. The postsequencing analysis was automated with a workflow involving UGENE, which also verified the complete conversion of cytosines to thymines for each clone in order to avoid false positives from partial bisulfite treatment.
Fig 2.
Microdissection of laryngeal papillomas. A representative sample of paraffin-embedded tissues shows, on the left, the hematoxylin and eosin-stained first slice and, in the middle and on the right, respectively, the second slice used for laser capture microdissection, before and after microdissection.
Fig 3.
Schematic illustration of the HPV6 URR. The gray boxes represent the PCR products obtained after bisulfite treatment. All the CpGs (black lines) present in the URR were included. Numbering is according to the amended version of HPV6b (9). The p97 promoter is indicated with a yellow arrow, and primers are indicated with orange arrows. E2BS, E2 binding site; NFI, nuclear factor I.
Even though we analyzed 360 clones, no methylated CpG was detected in any of the HPV6 URRs analyzed, although methylation was found in the URR of HPV16 from the control CaSki cells. The lack of methylation in HPV6 is in agreement with a recent study demonstrating the absence of methylation in the URR of HPV11 from a single patient with laryngeal papillomas (8). Nevertheless, the lack of methylation within our series of HPV6 URRs was unexpected, since a high frequency of hypermethylation has been observed in host tumor suppressor genes in laryngeal papillomas (25). However, HPV6 has also been detected in nondiseased respiratory sites (17, 22), and it is tempting to speculate that expression of HPV6 is repressed by methylation in such tissues. In lesions characterized by koilocytes, a hallmark of permissive HPV infections, it has been shown that HPV16 URR methylation varies depending on the degree of differentiation of the squamous epithelium (28). In that study, the central segment (enhancer) was methylated in basal cells and gradually decreased toward the upper cells. The most significant difference was observed in the promoter segment, which was intensively methylated in the superficial cells while it was totally unmethylated in the basal and intermediate cells. In contrast, our study of HPV6 in papillomas with koilocytes (Fig. 2) revealed that the HPV6 region is completely unmethylated in the various segments of the URR, regardless of the differentiation status of the squamous cells.
It is also important to note that unlike HPV16, HPV6 remains episomal. Among our samples, we had a detection rate of 100% for the 8-kb HPV6 amplicon by long-range PCR, indicating a high copy number of episomal HPV6. Therefore, the lack of methylation in the URR is probably representative of episomal HPV6 in laryngeal papillomas. To the best of our knowledge, there is no study that has demonstrated HPV6 integration among benign laryngeal papillomas, but we cannot completely exclude the possibility that integrated HPV6 can be methylated in these lesions. Nonetheless, the episomal form of HPV6 among our samples cannot explain the lack of methylation, as the episomal form of HPV16 has been shown to be methylated (2, 10, 14).
To our knowledge, this represents the first study of the methylation status of a series of low-risk HPV isolates. However, our exploratory study has limitations due to the small number of HPV6 isolates characterized and because we restricted the methylation analysis to the URRs of aerodigestive tract papillomas. In future studies, it would be interesting to investigate the methylation status of HPV6 also in genital condyloma, as well as methylation in the L1 region of HPV6, since the L1 region has demonstrated increased methylation in HPV16 from precancerous and cancerous cervix lesions (26). Further studies are also required to establish whether general and distinctive patterns of methylation exist among high- and low-risk HPV types.
In summary, we have demonstrated for the first time that the URR of HPV6 is completely unmethylated in papillomas of the aerodigestive tract, regardless of epithelial differentiation status.
Nucleotide sequence accession numbers.
All the sequences represented novel variants and were deposited in GenBank under accession numbers JN252314 to JN252323.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Cancer Society and Skåne Regional Research Funds (ALF).
We are grateful to Katarina Olofsson, Department of Otorhinolaryngology, Umeå, Sweden, for kindly sharing samples, and to the laryngologists who actively participated in the collection of samples. We thank Elise Nilsson for the preparation of histological cuts and Martin E. Johansson for helpful advice and discussion. Also, we thank Cecilia Wahlström for technical assistance.
Ethical approval for this study was granted by the Ethical Committee of Lund University (473/2006).
Footnotes
Published ahead of print 3 October 2012
REFERENCES
- 1. Bernard H-U, et al. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhattacharjee B, Sengupta S. 2006. CpG methylation of HPV 16 LCR at E2 binding site proximal to p97 is associated with cervical cancer in presence of intact E2. Virology 354:280–285 [DOI] [PubMed] [Google Scholar]
- 3. De-Castro Arce J, Göckel-Krzikalla E, Rösl F. 2012. Silencing of multi-copy HPV16 by viral self-methylation and chromatin occlusion: a model for epigenetic virus-host interaction. Hum. Mol. Genet. 21:1693–1705 [DOI] [PubMed] [Google Scholar]
- 4. de Villiers EM, Gissmann L, zur Hausen H. 1981. Molecular cloning of viral DNA from human genital warts. J. Virol. 40:932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding D-C, et al. 2009. Methylation of the long control region of HPV16 is related to the severity of cervical neoplasia. Eur. J. Obstet. Gynecol. Reprod. Biol. 147:215–220 [DOI] [PubMed] [Google Scholar]
- 6. Donne AJ, Hampson L, Homer JJIN, Hampson IN. 2010. The role of HPV type in recurrent respiratory papillomatosis. Int. J. Pediatri. Otorhinolaryngol. 74:7–14 [DOI] [PubMed] [Google Scholar]
- 7. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gáll T, Kis A, Fehér E, Gergely L, Szarka K. 2011. Virological failure of intralesional cidofovir therapy in recurrent respiratory papillomatosis is not associated with genetic or epigenetic changes of HPV11: complete genome comparison of sequential isolates. Antiviral Res. 92:356–358 [DOI] [PubMed] [Google Scholar]
- 9. Heinzel P, et al. 1995. Variation of human papillomavirus type 6 (HPV-6) and HPV-11 genomes sampled throughout the world. J. Clin. Microbiol. 33:1746–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kalantari M, Chase DM, Tewari KS, Bernard H-U. 2010. Recombination of human papillomavirus-16 and host DNA in exfoliated cervical cells: a pilot study of L1 gene methylation and chromosomal integration as biomarkers of carcinogenic progression. J. Med. Virol. 82:311–320 [DOI] [PubMed] [Google Scholar]
- 11. Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. 2003. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 77:12450–12459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kocjan BJ, et al. 2009. Prevaccination genomic diversity of human papillomavirus genotype 6 (HPV 6). Virology 391:274–283 [DOI] [PubMed] [Google Scholar]
- 13. Larson DA, Derkay CS. 2010. Epidemiology of recurrent respiratory papillomatosis. APMIS 118:450–454 [DOI] [PubMed] [Google Scholar]
- 14. Mazumder Indra D, et al. 2011. Genetic and epigenetic changes of HPV16 in cervical cancer differentially regulate E6/E7 expression and associate with disease progression. Gynecol. Oncol. 123:597–604 [DOI] [PubMed] [Google Scholar]
- 15. Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167 [DOI] [PubMed] [Google Scholar]
- 16. Park I-S, et al. 2011. Characterization of the methylation patterns in human papillomavirus type 16 viral DNA in head and neck cancers. Cancer Prev. Res. (Philadelphia). 4:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pignatari S, Smith EM, Gray SD, Shive C, Turek LP. 1992. Detection of human papillomavirus infection in diseased and nondiseased sites of the respiratory tract in recurrent respiratory papillomatosis patients by DNA hybridization. Ann. Otol. Rhinol. Laryngol. 101:408–412 [DOI] [PubMed] [Google Scholar]
- 18. Rambaut A. 2009. FigTree v1.3. http://beast.bio.ed.ac.uk/figtree
- 19. Schmitt M, et al. 2006. Bead-based multiplex genotyping of human papillomaviruses. J. Clin. Microbiol. 44:504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwarz E, et al. 1983. DNA sequence and genome organization of genital human papillomavirus type 6b. EMBO J. 2:2341–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silvestro D, Michalak I. 2011. raxmlGUI: a graphical front-end for RAxML. Organ. Divers. Evol. doi:10.1007/s13127-011-0056-0 [Google Scholar]
- 22. Smith EM, Pignatari SS, Gray SD, Haugen TH, Turek LP. 1993. Human papillomavirus infection in papillomas and nondiseased respiratory sites of patients with recurrent respiratory papillomatosis using the polymerase chain reaction. Arch. Otolaryngol. Head Neck Surg. 119:554–557 [DOI] [PubMed] [Google Scholar]
- 23. Söderlund-Strand A, Carlson J, Dillner J. 2009. Modified general primer PCR system for sensitive detection of multiple types of oncogenic human papillomavirus. J. Clin. Microbiol. 47:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics (Oxford) 21:456–463 [DOI] [PubMed] [Google Scholar]
- 25. Stephen JK, et al. 2010. Consistent DNA hypermethylation patterns in laryngeal papillomas. Int. J. Head Neck Surg. 1:69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun C, Reimers LL, Burk RD. 2011. Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer. Gynecol. Oncol. 121:59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ure A, et al. 2011. Characterization of the complete genomes of Camelus dromedarius papillomavirus types 1 and 2. J. Gen. Virol. 92:1769–1777 [DOI] [PubMed] [Google Scholar]
- 28. Vinokurova S, von Knebel Doeberitz M. 2011. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS One 6:e24451 doi:10.1371/journal.pone.0024451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342–350 [DOI] [PubMed] [Google Scholar]