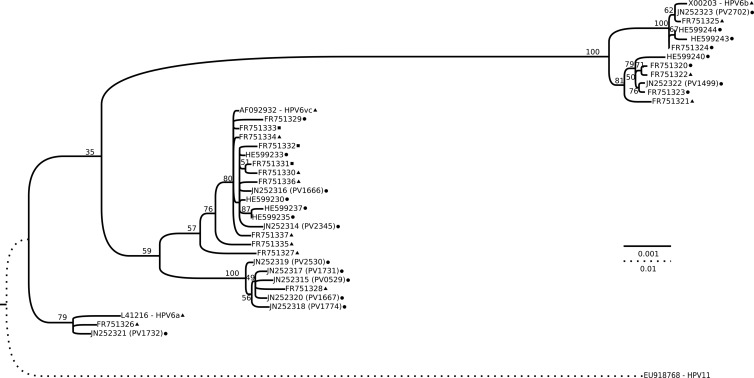
Fig 1.
Phylogenetic tree based on the complete genomes of all unique HPV6 variants available in GenBank. All accession numbers are indicated in the leaves of the tree, and for our 10 new HPV6 genomes the isolate name is given in parentheses. The isolation source is indicated, as follows: circle, laryngeal papilloma; triangle, condyloma acuminata; square, both laryngeal papilloma and condyloma acuminata (for genomes deposited in GenBank with identical sequences but different isolation sites). The tree was obtained by the maximum likelihood approach using RAxML software and rooted with HPV11. Bootstrap (800) support values are indicated in each branch as percentages. The bars indicate the number of substitutions per site for the two different scales used.