Figure 3. Mutagenesis of mycobacterial genomes.
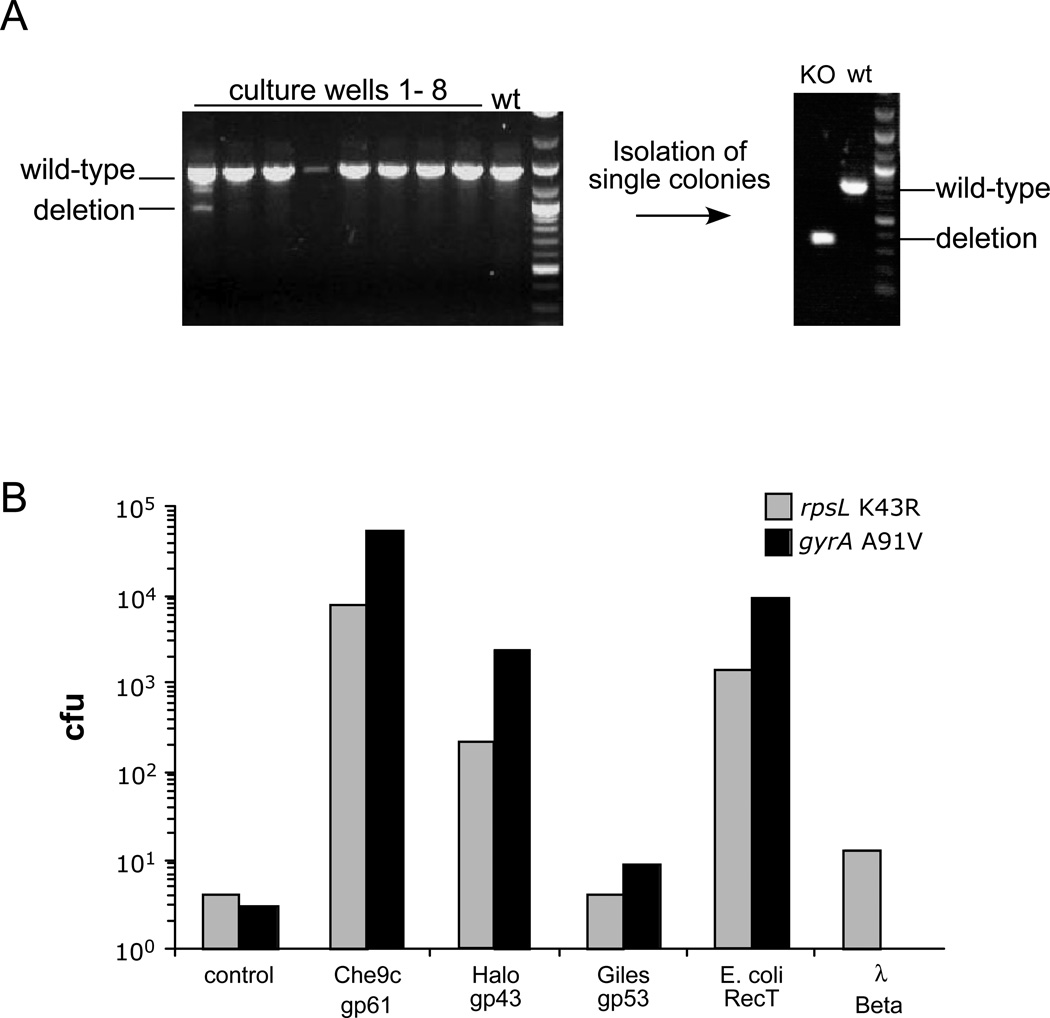
A. Constructing an unmarked deletion of the M. smegmatis leuD gene using recombineering. Induced electrocompetent M. smegmatis/pJV76amber cells (pJV76amber is a derivative of pJV53 containing a mutant HygR gene) were co-electroporated with a 200 bp leuD deletion substrate along with an oligonucleotide that repairs the mutant plasmid HygR gene. After dilution to approximately 10 cells per sample, hygromycin selection and growth, at least one sample (#1) contained the mutant allele, and a leu− strain was readily recovered and verified by PCR.
B. Recombinases (SSAPs) were assayed for ssDNA recombineering activity in M. smegmatis by their ability to introduce streptomycin-resistant (rpsL K43R) and ofloxacin-resistant (gyrA A91V) mutations. SSAP genes are expressed from the acetamidase promoter and translationally-fused to the pLAM12 vector48 rather than using their native translation initiation signals49. The number of drug resistant colony forming units (cfu) obtained for each drug target are shown.