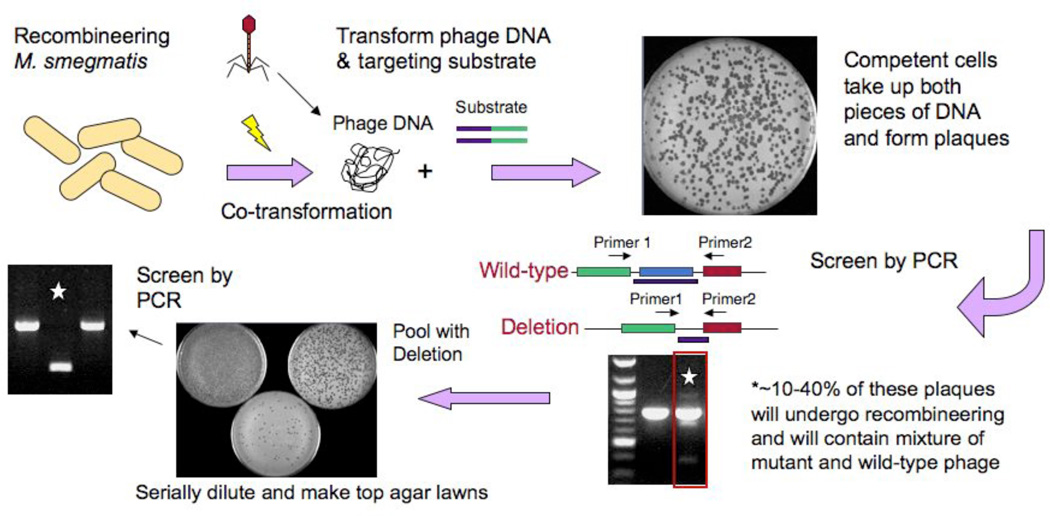
Figure 4. Mycobacteriophage recombineering.
Schematic illustration of the strategy utilized to adapt mycobacterial recombineering for use on mycobacteriophages. Induced electrocompetent M. smegmatis/pJV53 cells are co-electroporated with phage DNA (50–100 ng) and a 200bp dsDNA recombineering substrate (50–500 ng), mixed with M. smegmatis plating cells, and plated for individual plaques. Approximately 10–40% of plaques are mixed, containing both the mutant and wild-type alleles, and are distinguishable by PCR. A mixed plaque can be diluted and plated for analysis of individual plaques or for preparation of a lysate. If a non-essential gene is deleted then a mutant phage derivative can be readily identified by PCR, but if the gene is essential, then the mutant allele is no longer present in the lysate.