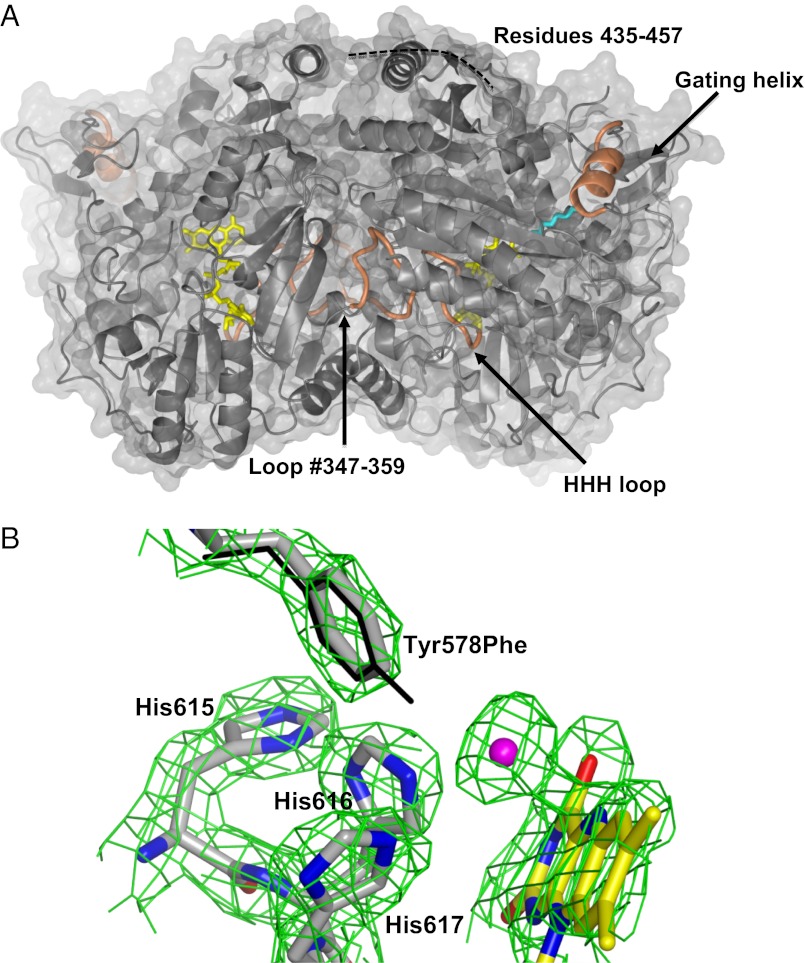
Fig. 2.
Crystal structure of ADPS. (A) The dimer with the molecular twofold axis vertical in the plane of the paper. The FAD is shown in yellow; the bound aliphatic ligand, in cyan. Loop 347–359, the “HHH loop” (615–622), and the gating α-helix (126–137) are shown in orange. # indicates that loop 347–359 is part of the active site of the twofold related subunit. ADPS is associated to the inner side of the peroxisomal membrane (13). This localization enables the liposoluble fatty alcohol substrate and fatty acid product to directly diffuse to and from the membrane (29). By analogy with the Dictyostelium ADPS structure, the site of membrane association is predicted to be on the upper side of the dimer, in the area embraced by the disordered 435–457 segment (14). (B) The final 2Fo-Fc–weighted electron density map (contoured at the 1.4 σ level) of the active site in the Tyr578Phe mutant. Protein carbons are shown in gray; flavin carbons, in yellow; oxygens, in red; nitrogens, in blue; and chloride, in magenta. The Tyr578 side chain of the WT protein is superimposed and shown with thin bonds. The only detectable change is the strong electron density peak in direct contact with the flavin N5 atom. We have interpreted this peak as a chloride ion bound to flavin, the negative charge of which is compensated for by the His side chains of the HHH loop. This observation supports the role of these His residues in stabilization of the negatively charged enolate form of the substrate (Fig. 1).