Abstract
Metazoan histone mRNAs are unique: their pre-mRNAs contain no introns and the mRNAs are not polyadenylated ending instead in a conserved stem-loop structure. In Drosophila, canonical poly(A) signals are located downstream of the normal cleavage site of each histone gene, and are utilized when histone 3’end formation is inhibited. Here we define a sub-complex of poly(A) factors required for histone pre-mRNA processing. We demonstrate that Symplekin, CPSF73 and CPSF100 are present in a stable complex and interact with histone specific processing factors. We use chromatin immunoprecipitation to show that Symplekin and CPSF73, but not CstF50, cotranscriptionally associate with histone genes. Depletion of SLBP recruits CstF50 to histone genes. Knockdown of CPSF160 or CstF64 downregulates Symplekin but does not affect histone pre-mRNA processing or association of Symplekin with the histone locus. These results suggest that a common core cleavage factor is required for processing of histone and polyadenylated pre-mRNAs.
Introduction
Metazoan histone mRNAs are unique among eukaryotic mRNAs in that they contain no introns and end in a conserved stem-loop structure rather than a poly(A) tail. Formation of a mature histone mRNA requires only a single endonucleolytic cleavage which is directed by two cis elements, a highly conserved stem loop (SL) 5’ of the cleavage site and a less conserved purine-rich histone downstream element (HDE) 3’ of the cleavage site. There are two histone specific RNA processing factors, the stem loop binding protein (SLBP) which binds the SL and the U7 RNA of the U7 small nuclear ribonucleoprotein (U7 snRNP) which base pairs with the HDE (Marzluff et al., 2008). The U7 snRNP consists of the U7 snRNA and a heteroheptameric ring of Sm proteins in which the spliceosomal Sm proteins SmD1 and SmD2 are replaced by Lsm10 and Lsm11 (Pillai et al., 2003). Cleavage is catalyzed by CPSF73 (Dominski et al., 2005b) and Symplekin has been implicated as the scaffold which coordinates formation of the cleavage complex (Kolev and Steitz, 2005). Following processing, the mature mRNA is escorted into the cytoplasm by SLBP (Sullivan et al., 2009) where SLBP participates in efficient translation of histone mRNA (Sanchez and Marzluff, 2002).
Cleavage and polyadenylation of all other metazoan mRNAs requires two multi-protein complexes termed the cleavage and polyadenylation specificity factor (CPSF) and the cleavage stimulation factor (CstF), which recognize signals upstream and downstream of the cleavage site, respectively. CPSF is composed of CPSF30, CPSF73, CPSF100 and CPSF160, which interact with one another [reviewed in (Mandel et al., 2008)] and with the AAUAAA polyadenylation signal that is recognized by CPSF160 (Keller et al., 1991; Murthy and Manley, 1995). Both CPSF73 and CPSF100 have putative β-lactamase domains, and CPSF73 has been described as the endonuclease for both poly(A) (Mandel et al., 2006) and histone mRNAs (Dominski et al., 2005b). CPSF100 has also been shown to play an important role in the cleavage reaction (Kolev et al., 2008) although it lacks critical residues required for catalysis. CstF64, a member of the CstF complex binds the downstream GU-rich element required for polyadenylation (Yoshio and Manley, 1997).
Symplekin was originally identified as a tight junction protein in mammalian cells (Keon et al., 1996) and its yeast homolog, Pta1p, was initially characterized as being essential for pre-tRNA processing (O’Connor and Peebles, 1992). Symplekin has subsequently been shown to interact with both CPSF and CstF in yeast (Preker et al., 1997; Zhao et al., 1999) and mammals (Takagaki and Manley, 2000; Vethantham et al., 2007). Additionally, Symplekin was defined as the heat labile factor (Gick et al., 1987) required for histone pre-mRNA processing (Kolev and Steitz, 2005).
In Drosophila, there are 100 tandem copies of a 5 kb repeat unit containing a single copy of each histone gene (H2A, H2B, H3, H4 and H1) on the long arm of chromosome 2. This histone gene cluster localizes to nuclear structures termed histone locus bodies (HLBs) which are located in close proximity to Cajal bodies (CBs) (Liu et al., 2006). HLBs also contain U7 snRNP, Symplekin (Wagner et al., 2007), and Mpm-2 (White et al., 2007), consistent with a role for these nuclear bodies in histone mRNA biosynthesis.
Various RNA processing events have been shown to occur cotranscriptionally [reviewed in (Bentley, 2005; Buratowski, 2005; Neugebauer, 2002; Proudfoot, 2004)] and some components of the poly(A) apparatus directly bind the CTD of RNA Pol II (Kyburz et al., 2003; McCracken et al., 1997; Phatnani and Greenleaf, 2004) suggesting that polyadenylation factors can be loaded onto the polymerase. Polyadenylation factors are also associated with genes encoding mRNAs as detected by chromatin immunoprecipitation (ChIP). Additionally, the CTD is important for efficient cleavage/polyadenylation reaction in vivo (Hirose and Manley, 1998). Finally, in vitro, Drosophila RNA Pol II pauses just 3’ of the processing site of histone genes, in a position that would allow cotranscriptional assembly of the processing complex (Adamson and Price, 2003). These data support the idea that the 3’ ends of both polyadenylated and histone mRNAs are formed cotranscriptionally.
In Drosophila the 3’ ends of four of the histone genes are less than 500 nts from the 3’ end of an adjacent gene (transcribed from the opposite strand, Fig. 1A). Thus, to prevent read-through into the adjacent gene, it is essential to efficiently terminate transcription. There are cryptic polyadenylation signals downstream of each Drosophila histone gene. If the processing efficiency of histone mRNAs is reduced either by mutation or knockdown of factors required for histone mRNA processing, then RNA Pol II reads through and the mRNAs become polyadenylated (Godfrey et al., 2006; Sullivan et al., 2001).
Figure 1. Knockdown of pre-mRNA processing factors results in misprocessed histone mRNA.
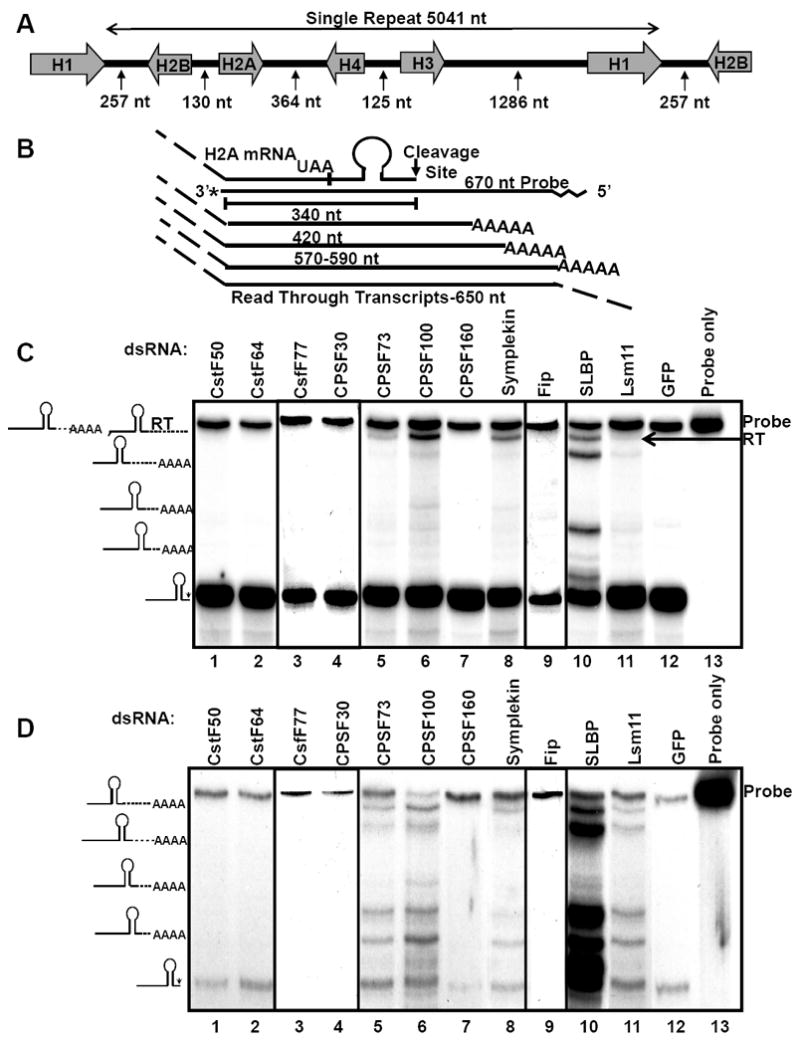
(A) A schematic of one tandem repeat of the histone gene locus is shown. The numbers indicate the distance between the 5’ or 3’ ends of the mRNAs . (B) Schematic of S1 nuclease protection assay. A 670 nt DNA fragment containing 650 nts of the 3’ end of H2A gene and 20 nts of plasmid sequence at the 5’ end was labeled with α-32P-dCTP on the 3’ end. The probe was hybridized to total cell RNA and digested with S1 nuclease. Properly processed H2A mRNA yields a protected fragment of 340 nt extending to the 3’ end of the mRNA while misprocessed transcripts give fragments ranging in size from 340-600nt. Any read-through protects a 650 nt fragment. Two gels were necessary to accommodate all samples in the same order in Figs. 1C, 1D and 2A, boundaries have been clearly indicated. Relative amounts of properly processed histone pre-mRNA from lanes 3, 4 and 9 were not significantly different from a control treated sample on the same gel. (C) Dmel-2 cells were treated with the indicated dsRNAs and total RNA was prepared. 12.5 μg of RNA was analyzed by S1 nuclease protection assay. Diagrams at left indicate RNA species corresponding to protected fragments. The undigested probe is shown in lane 13. Read-through transcripts (RT) are marked with an arrow. (D) 25 μg of total RNA was fractionated on oligo(dT) cellulose and the purified poly(A)+ RNA was used for S1 nuclease protection assay as in (B). A small amount of the processed histone mRNA nonspecifically binds to the oligo(dT) cellulose.
A recent RNA interference screen in Drosophila implicated a subset of polyadenylation factors, Symplekin, CPSF73 and CPSF100 in histone pre-mRNA processing, while other polyadenylation factors did not score in the screen (Wagner et al., 2007). To further investigate the role of these proteins in histone pre-mRNA processing, we first examined the effect of RNAi-depletion of these factors on the 3’ end of endogenous histone mRNA. We carried out co-immunoprecipitation (coIP) and ChIP experiments to demonstrate that Symplekin, CPSF-73 and CPSF-100 are part of a core cleavage factor involved in cotranscriptional histone mRNA 3’ end processing.
Results
The histone genes in Drosophila are clustered in a tandemly repeated unit containing one copy of each of the five genes. The number of nucleotides separating each gene is small (Fig. 1A); thus, efficient processing and transcription termination are required to prevent transcription into neighboring ORFs. To ensure production of histone mRNAs, multiple Drosophila species have evolved canonical poly(A) sites downstream of the normal cleavage site of each histone gene. Mutation of histone processing factors such as SLBP (Lanzotti et al., 2002) or components of the U7 snRNP (Godfrey et al., 2006) results in the expression of polyadenylated histone mRNAs from each of the five histone genes. The presence of these polyadenylated mRNAs indicates that histone 3’ end processing is inefficient, allowing us to address the role of protein factors in this process.
A subset of poly(A) factors is required for histone pre-mRNA processing in vivo
CPSF73, CPSF100 and Symplekin were previously shown to activate a histone misprocessing reporter (Wagner et al., 2007) but the effect of their knockdown on expression of endogenous histone mRNAs was not examined. To study Drosophila histone pre-mRNA 3’ end processing in vivo, we RNAi depleted both histone and general poly(A) processing components in Dmel-2 cells, and performed an S1 nuclease protection assay to assess the processing of histone H2A mRNA (Fig. 1). Western and Northern blotting were used to assay the levels of processing factors (Fig. 2A) and their mRNAs (Fig. S1). Dmel-2 cells were bathed in dsRNAs targeting the transcripts of 3’ end processing factors for five days and RNA and nuclear extracts (NEs) were then prepared from the cells. Total RNA was hybridized to a 670 nt probe. The 3’ 340 nts of this probe base pair to the 5’ end (including the processing site) of H2A. The probe also contains 310 nts complementary to the 3’ flanking sequence which hybridized to polyadenylated histone H2A mRNAs (Lanzotti et al., 2002). 20 nts of non-histone sequence were added to the 5’ end of the probe allowing us to distinguish undigested probe from transcripts that went beyond the end of the probe (referred to as read-through transcripts). Hybridization of the probe was followed by digestion with S1 nuclease to map the 3’ end of the RNA transcripts. Separation of the protected fragments by gel electrophoresis reveals properly processed histone mRNA as well as longer discrete fragments in samples depleted of various 3’ end processing factors (Fig. 1C). Properly processed H2A mRNA yields a protected fragment of 340 nts, while misprocessed transcripts utilizing one of the downstream poly(A) sites resulted in several larger protected species between 340 and 650 nts (Fig. 1C, compare lanes 10 and 12).
Figure 2. The core cleavage factor interacts with histone specific processing factors.
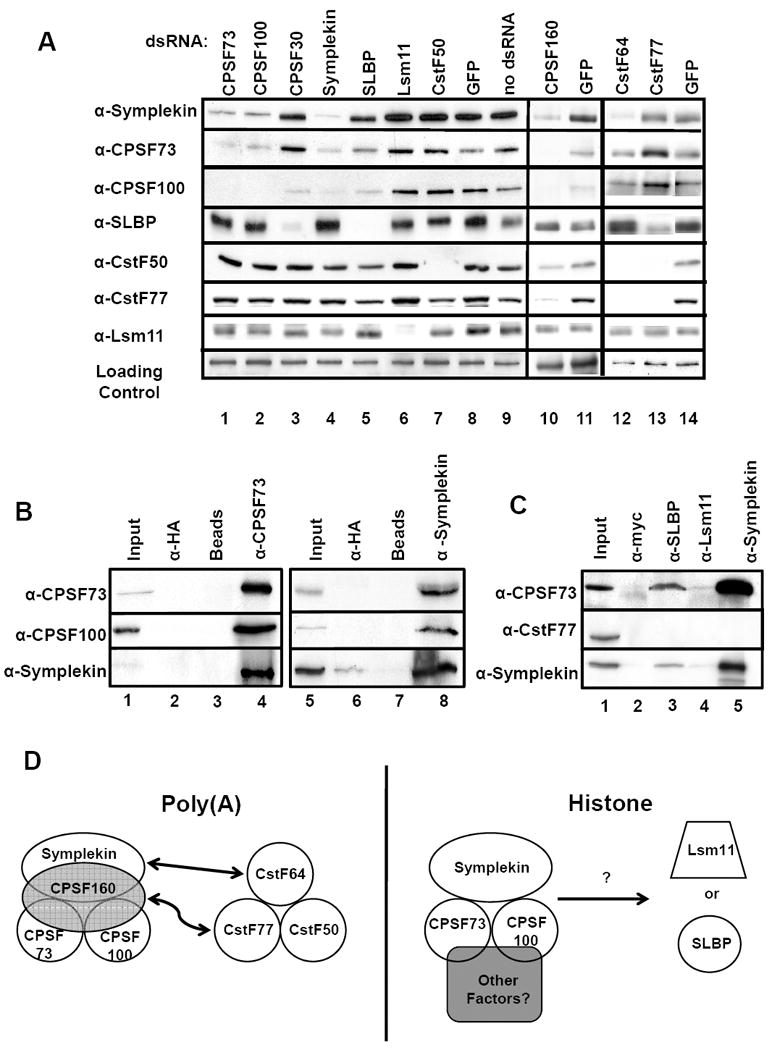
(A) The indicated proteins were RNAi-depleted and NEs were prepared. 15 μg of NE was resolved by SDS-PAGE and western blots were performed using indicated antibodies. The loading control is a cross-reacting band from the Symplekin blot. (B) Dmel-2 NEs were used in IP experiments with indicated antibodies under high detergent conditions. α-HA and beads alone IPs were included as controls. Inputs represent 2.5% of starting material. (C) IPs were performed as in A except under low detergent conditions. (D) Diagram of core cleavage factor interactions with poly(A) and histone specific RNA processing factors.
Depletion of the histone specific pre-mRNA processing factor SLBP resulted in substantial misprocessing in vivo and multiple polyadenylated mRNA species (Fig. 1C, lane 10) as previously described for SLBP mutant embryos (Lanzotti et al., 2002) and larvae (Godfrey et al., 2006). Even when SLBP (and other factors) is depleted, the majority of the histone mRNA is still properly processed, indicating that the residual levels of the factors were sufficient for processing much of the histone mRNA, since in null mutants of SLBP or U7 snRNP only polyadenylated mRNA is produced (Godfrey et al., 2006).
In agreement with the results obtained with the reporter, knockdown of some components of the CPSF complex (CPSF73 or CPSF100) caused histone pre-mRNA misprocessing while knockdown of CPSF30 or CPSF160 had no effect on these transcripts (Fig. 1C, lanes 4-7). Symplekin depletion also resulted in H2A misprocessing (Fig. 1C, lane 8), while Fip knockdown had no detectable effect on endogenous H2A mRNA (Fig. 1C lane 9) though it scored weakly on the misprocessing reporter (Wagner et al., 2007). In contrast, depletion of CstF complex components had no effect on histone 3’ end formation (Fig. 1C lanes 1-3). Interestingly, knockdown of polyadenylation factors primarily resulted in production of the long read-through product (Fig. 1C, lanes 5, 6, 8), unlike the specific mRNAs produced in knockdown of SLBP and Lsm11.
To confirm that misprocessed H2A mRNA was polyadenylated, total RNA was fractionated on oligo d(T) cellulose to select poly(A)+ RNA prior to the S1 nuclease protection assay. Enrichment of poly(A)+ mRNA reveals that depletion of both histone specific and general poly(A) factors results in a similar pattern of polyadenylated histone mRNAs in each sample. (Fig. 1D, lanes 5-8, 10-11). However, the pattern of protected histone H2A mRNA fragments in total RNA and poly(A)+ RNA of the CPSF73, CPSF100 and Symplekin knockdowns (compare Figs. 1C and 1D, lanes 5, 6, 8) were strikingly different. This was not the case for SLBP and Lsm11 knockdowns (Figs. 1C and 1D, lanes 10 and 11). Most of the longest transcripts detected when polyadenylation factors were depleted were not selected on d(T) cellulose and hence were not polyadenylated. These results indicate that knockdown of CPSF73, CPSF100 and Symplekin causes read-through of both the canonical histone mRNA 3’ end processing signals and the additional cryptic polyadenylation sequences and are consistent with a defect in polyadenylation as well as histone pre-mRNA processing. In contrast, depletion of histone specific factors SLBP and Lsm11 resulted in similar ratios of polyadenylated H2A mRNA in both total and poly(A)+ RNA. This suggests that there is little if any accumulated read-through, unprocessed mRNA in these samples.
These data indicate that a subset of polyadenylation factors (CPSF73, CPSF100 and Symplekin) is required for both histone and polyadenylated pre-mRNA 3’ end formation in vivo. Depletion of these factors results in accumulation not only of misprocessed polyadenylated histone mRNAs, as is seen in knockdowns of histone specific pre-mRNA processing factors, but also of unprocessed read-through transcripts, due to reduction in efficiency of polyadenylation.
Components of the core cleavage factor are codepleted by RNAi in vivo
Before conducting S1 nuclease assays on RNAi-depleted samples, western blots were performed (with available antibodies) to determine the knockdown efficiency of poly(A) and histone processing factors. Strikingly, depletion of some factors caused codepletion of other components, suggesting that these proteins complex with one another. Similar results have been observed for homologous polyadenylation factors in yeast (Kyburz et al., 2003; Zhao et al., 1999). Depletion of CPSF73, CPSF100 or CPSF160 results in codepletion of CPSF73, CPSF100 and Symplekin, while CSPF30 depletion had only a mild effect on the level of CPSF100 (Fig. 2A, lanes 1-3 and 10). Additionally, knockdown of Symplekin also caused codepletion of CPSF73 and CPSF100 (Fig. 2A lane 4). Depletion of the CstF subunits CstF64 and CstF77, resulted in codepletion of CstF50 (Fig. 2A, lanes 12 and 13), although knockdown of CstF50 showed only modest codepletion of CstF77 (Fig. 2A, lane 7). There was no effect on SLBP and Lsm11 levels with RNAi-depletion of polyadenylation factors with the lone exception of a variable SLBP depletion in CPSF30 knockdowns. This depletion was not significant enough to affect endogenous histone mRNA processing (Figure 1C, lane 4) and was not investigated further. Knockdown of SLBP had a minor affect on CPSF100 level while Lsm11 knockdown did not affect any other factors tested (Fig. 2A, lanes 5-6).
Notably, while knockdown of CPSF160 reduced the levels of CPSF73, CPSF100 and Symplekin, CPSF160 depletion did not have an effect on histone pre-mRNA processing (Figs. 1C and 1D, lane 7). Similarly, knockdown of CstF64 resulted in codepletion of Symplekin (Fig. 2A, lane 12), but also did not affect histone pre-mRNA processing (Figs. 1C and 1D, lane 2). None of the knockdowns significantly affected mRNA levels of other factors, indicating that the codepletions are not due to the reduction of levels of specific mRNAs (Figure S1).
These data indicate that components of both the CPSF and CstF complexes can affect the stability of proteins within their respective complexes. Additionally, Symplekin protein levels are reduced by knockdown of the CPSF components CPSF73, CPSF100 and CPSF160 as well as CstF64 but not the other CstF subunits.
CPSF73, CPSF100 and Symplekin form a stable complex and also interact with histone mRNA specific processing factors
To directly test for protein-protein interactions among Drosophila 3’ end processing factors, coIP experiments were performed with NEs from Dmel-2 cells. We prepared polyclonal CPSF73 and Symplekin antibodies which were effective for IP, and used these to characterize the interactions between endogenous Symplekin and members of the CPSF complex. CPSF100 and Symplekin coIP with CPSF73 under stringent conditions (50 mM Tris, pH 8.0, 1% NP40, 0.25% deoxycholate, 150 mM NaCl; Fig. 2B, lane 4). This result was confirmed by coIP of CPSF73 and CPSF100 with the α-Symplekin antibody (Fig. 2B, lane 8). The high efficiency of coIP of these three endogenous factors strongly supports the idea that CPSF73, CPSF100 and Symplekin are part of a stable complex(es) in vivo.
To determine if components of the cleavage factor interact with specific histone pre-mRNA processing factors, coIP experiments with α-SLBP and α-Lsm11 antibodies were performed using NE from Dmel-2 cells. CPSF73 coIPs with both SLBP and Lsm11 (Fig. 2C, lanes 3-5) under less stringent conditions (50mM Tris, pH 8.0, 0.1% NP-40, 150 mM NaCl), although neither interaction is as strong as that between CPSF73 and Symplekin as these interactions were undetectable under stringent conditions (data not shown). Symplekin also coIPs with SLBP and, to a lesser extent, Lsm11 (Fig. 2C, lanes 3-5). While a large amount of the Symplekin, CPSF73 and CPSF100 coimmunoprecipitated with one another, only a small amount of the CPSF73 or Symplekin precipitated with SLBP or Lsm11. CstF77 did not coIP with Symplekin, CPSF73, Lsm11 or SLBP, consistent with depletion of CstF77 not having an effect on the concentrations of these proteins. These interactions are not RNA-dependent as neither RNAse treatment of the IPs nor actinomycin D treatment of the cells prior to nuclear extraction prevented coIP (data not shown).
Collectively, these data point to a core complex of cleavage factor components consisting of CPSF73, CPSF100 and Symplekin which interact very strongly with one another. This core complex may then bind different accessory proteins depending on the type of pre-mRNA to be processed. A schematic of possible interactions is shown in Figure 2D, indicating how a common core of CPSF73, CPSF100 and Symplekin might interact to form two different cleavage factors for processing of poly(A) mRNAs (Fig. 2D, left) versus histone mRNAs (Fig. 2D, right).
Components of the cleavage factor associate with histone genes in vivo
RNA processing factors have been shown to associate with genes via ChIP (Komarnitsky et al., 2000; Licatalosi et al., 2002; Proudfoot, 2004), either as complexes bound to the nascent RNA or by direct association with RNA Pol II. We investigated whether any of these factors associate with the Drosophila histone genes in vivo. Primer sets located within the coding regions of H2A, H2B, H3 and H4, the promoter regions of H2A/H2B and H3/H4 and a region downstream of H3 were used to amplify regions of the ~5kb histone gene cluster (Fig. 3A). There are multiple polyadenylation sites in the 1.3 kb intergenic region between H3 and H1 that are only used when histone mRNA processing is inhibited (Lanzotti et al., 2002). It should be noted that the small size of the histone genes (~400 bp) approaches the limit of the resolution of the chromatin shearing required for the ChIP assay (200-500 bp fragments) and thus differences between the 5’ and 3’ ends of these genes may not be readily distinguishable. As a control for the association of canonical poly(A) factors with a gene encoding a polyadenylated mRNA, primers were also designed to exon 6 and the first of two poly(A) sites of the domino gene (Fig. 3A). In contrast to the histone genes, these probes are well separated.
Figure 3. RNA processing factors are recruited to histone genes in vivo.
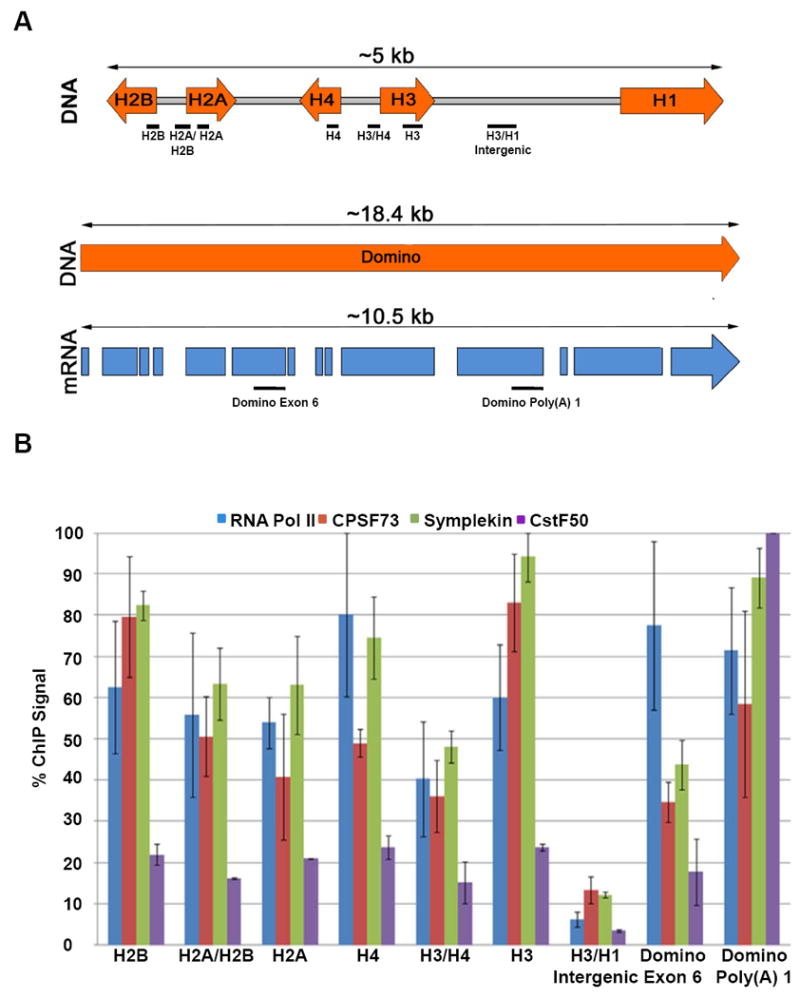
(A) Diagram of ChIP assay design. The oligo sets correspond to the coding regions of H2A, H2B, H3 and H4, the promoters of the H2A/H2B and H3/H4 pairs and the H3/H1 intergenic region. Control oligo sets were designed to exon 6 and the proximal poly(A) site of the domino gene. (B) ChIP assays were performed using antibodies to the Rpb3 subunit of RNA polymerase II, CPSF73, Symplekin and CstF50. Columns represent average relative ChIP signal normalized to the maximal value for each antibody. Data are presented as the average of 3 independent experiments ± SD.
ChIP experiments were performed with antibodies to CPSF73, CstF50, RNA Pol II (Rpb3) and Symplekin. The relative amounts of DNA associated with each fragment were quantified using qPCR and SYBRGreen fluorescence. Using an antibody which recognizes all forms of RNA Pol II without preference for phosphorylation state of the CTD, we found that RNA Pol II associated with all portions of the histone genes and the domino gene. Within the promoter and coding regions of each of the genes, the polymerase occupancy was between 40% and 80% of maximum whereas there was no RNA Pol II present in the intergenic region between the H3 and H1 genes (Fig. 3B, blue bars).
We next examined the localization of two components of the cleavage factor, CPSF73 and Symplekin. The domino gene showed an increased pattern of recruitment of both factors at the 3’ end of the gene relative to exon 6 (Figure 3B, red and green bars). Both CPSF73 and Symplekin were localized throughout the histone gene cluster with peak levels similar to those found at the 3’ end of the domino gene. Again, essentially no Symplekin or CPSF73 was detected in the intergenic between histone H3 and histone H1 in control cells. As a control, we carried out ChIP analysis with CstF50, a factor which has no role in histone pre-mRNA processing. CstF50 showed strong localization to the 3’ end of the domino gene and a much lower level of association with the histone genes than CPSF73 or Symplekin. Not surprisingly, there is little localization of CstF50 with the H3/H1 intergenic region (Fig. 3B, purple bars). Thus CPSF73 and Symplekin, factors involved in histone mRNA maturation, displayed cotranscriptional recruitment to histone genes as well as a canonical poly(A) gene.
SLBP knockdown causes recruitment of additional poly(A) factors to histone genes
Since knockdown of SLBP resulted in misprocessing and polyadenylation of histone mRNAs (Fig. 1C, lane10) we investigated how recruitment of processing machinery to the histone genes was affected by depletion of SLBP. IPs were performed from SLBP knockdown cells as in previous experiments. In these experiments we focused on the H3 gene using the primer sets which correspond to the promoter region (primer set H3/H4), the 3’ end of the coding region (primer set H3) and the H3/H1 intergenic region which contains two distal poly(A) sites (primer set H3/H1 intergenic). The domino poly(A) site was used as a control. RNA Pol II dropped from ~60% maximal occupancy at the 3’ end of histone H3 genes in control cells to ~40% in this same region in SLBP knockdown cells (Fig. 4A, top left). Also, RNA Pol II occupancy in the intergenic region increased from less than 10% in control cells to >40% in SLBP knockdown cells. CPSF73 and Symplekin showed similar recruitment patterns at the promoter and 3’ end in control and SLBP knockdown samples, however, both of these proteins also displayed a dramatic increase in ChIP signal in the intergenic region in SLBP knockdown cells (compare Fig. 4A, top right and bottom left). The most remarkable difference between control and SLBP knockdown cells was the enrichment of CstF50 across the histone H3 gene (Fig. 4A, bottom right). In control cells, this protein was recruited at only a low level, however, in cells depleted of SLBP it showed occupancy of >50% of maximum at the 3’ end and intergenic regions. CstF50 occupancy in the intergenic region increased from ~5% to ~70% in SLBP knockdown cells. This is consistent with recruitment of CstF50 to histone genes as part of the polyadenylation of histone pre-mRNAs in the absence of SLBP.
Figure 4. Loss of a histone specific processing factor results in recruitment of additional poly(A) factors to histone genes.
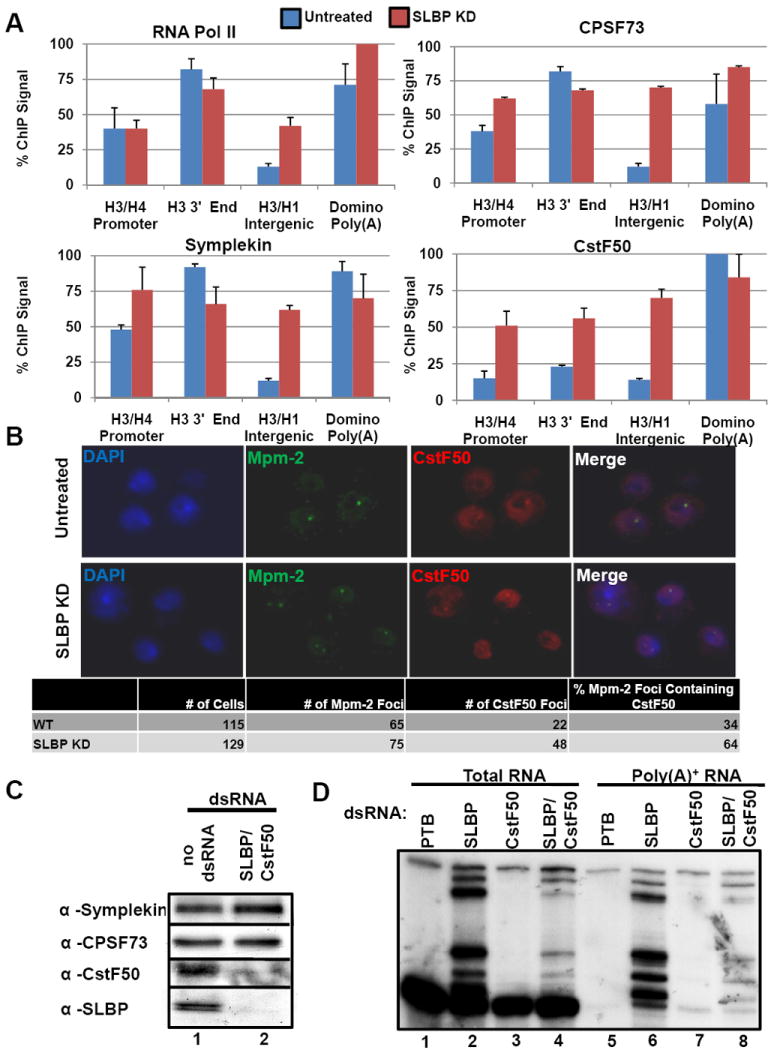
(A) ChIP assays were performed using lysates prepared from Dmel-2 cells treated with control (blue) or SLBP (red) dsRNAs. Columns represent the average relative ChIP signal normalized to the maximal value for each antibody. Data are presented as the average of 3 independent experiments +SD. Blue bars are untreated cells; red bars are values for SLBP knockdown cells. (B) Dmel-2 cells treated with control dsRNAs (top) or SLBP dsRNAs (bottom) were fixed and probed for CstF50 (green) and Mpm-2 (red), and also stained with DAPI (blue). The number of cells with Mpm-2 foci, and the number of cells with Mpm-2 foci that also contained CstF50 foci were counted. (C) Western blots of Dmel-2 cells treated with PTB dsRNA (control) or dsRNA targeting SLBP and CstF50. (D) Total RNA was prepared from cells treated as in (C) or with dsRNAs against SLBP or CstF50. S1 nuclease protection assays were performed on total RNA (lanes 1-4) or poly(A)+ RNA (lanes 5-8) as in Fig. 1 C, D.
Immunofluorescence (IF) experiments also showed increased recruitment of CstF50 to HLBs when SLBP is RNAi-depleted (Figure 4B). Cells were analyzed by IF for Mpm-2, an antigen that localizes to HLBs (White et al., 2007) and hence histone genes in S-phase Drosophila cells (>50% of cells), and CstF50. CstF50 was generally localized throughout the nucleoplasm in control cells, consistent with a role in processing a large number of genes. Foci of CstF50 were clearly visible in some of the SLBP knockdown cells and in 64% of the cells these foci colocalized with Mpm-2, consistent with them being in the HLB. In control cells, CstF50 foci were generally not visible, although in 34% of cells there were significant amounts of CstF50 that overlapped in localization with Mpm-2 (Fig. 4B). Thus SLBP knockdown results in an increased association of CstF50 with the HLB.
To determine the effect of simultaneously depleting both a histone and a poly(A) factor on histone pre-mRNA processing, protein lysates and RNA were made from Dmel-2 cells depleted of both SLBP and CstF50. Efficient knockdown of both proteins in the double knockdown was confirmed by western blot (Fig. 4C). An S1 nuclease protection assay was then performed on the RNA from the double knockdown as well as on RNA from SLBP and CstF50 individual knockdowns. SLBP knockdown again resulted in substantial misprocessing and polyadenylation (Fig. 4D, lanes 2 and 6) while depletion of CstF50 had no effect on histone pre-mRNA processing (Fig. 4D, lanes 3 and 7). The double knockdown, however, showed an intermediate phenotype in which the distal poly(A) site is preferentially used for polyadenylation (Fig. 4D, lanes 4 and 8), consistent with both histone pre-mRNA processing being affected by SLBP depletion and polyadenylation being affected by CstF50 depletion. Collectively, the data in Figure 4 demonstrate a role for the CstF complex in polyadenylation of misprocessed histone mRNAs and that CstF50 is not required for normal processing.
Depletion of Symplekin by RNAi-depletion of CPSF160 or CstF64 does not affect recruitment of the cleavage factor to histone genes
Knockdown of CstF64 or CPSF160 resulted in codepletion of factors required for histone pre-mRNA processing yet did not result in polyadenylation of histone mRNAs (Fig. 1C, lanes 2 and 7). Knockdown of CPSF160 codepletes CPSF73, CPSF100 and to a lesser extent, Symplekin, while CstF64 knockdown resulted in depletion of Symplekin (Fig. 2A, lanes 10 and 12). To determine whether this codepletion affects association of common cleavage factor components with either histone or poly(A) genes, we knocked down CstF64 or CPSF160 and performed ChIP. When CstF64 was depleted, RNA Pol II and CPSF73 showed maximum occupancy at the 3’ end of the H3 gene with no significant association in the intergenic region between H3 and H1. Additionally, CPSF73 and RNA Pol II had ~ 60% of maximal association with the promoter region of H3 and the proximal domino poly(A) site (Fig. 5A, top). Little difference was seen between the ChIP profiles of RNA Pol II and CPSF73 in the CstF64 knockdown samples as compared to the untreated control. Symplekin showed a similar pattern of association with the histone gene as both RNA Pol II and CPSF73, although there was a substantial decrease in the relative amount of Symplekin associated with the domino poly(A) site (Fig. 5A, bottom). This striking result is consistent with CstF64 knockdown causing codepletion of the Symplekin required for processing of poly(A) mRNAs, but had no effect on the Symplekin required for histone mRNA processing.
Figure 5. Knockdown of poly(A) specific factors does not affect recruitment of core cleavage factor to histone genes.
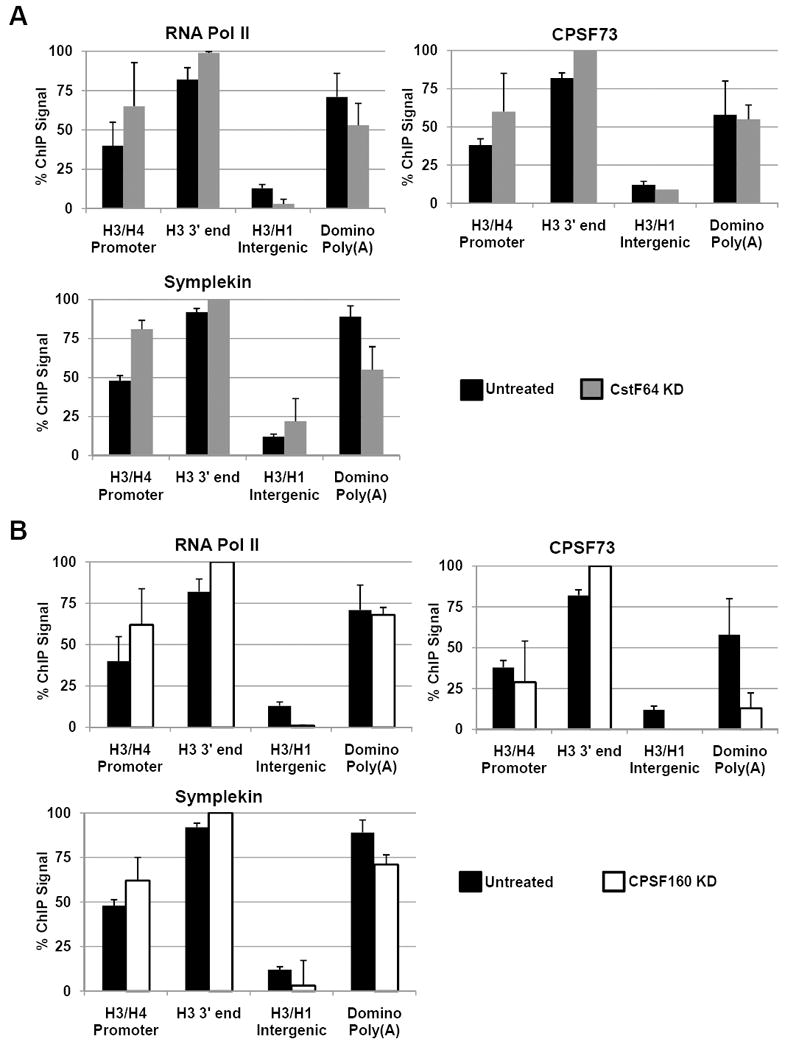
ChIP assays were performed as in Fig. 3B on (A) CstF64 or (B) CPSF160 knockdown cells. Columns represent the average relative ChIP signal normalized to the maximal value for each antibody. Data are presented as the average of 3 independent experiments +SD.
We next examined the effect of CPSF160 knockdown on association of RNA Pol II and RNA processing factors with the histone H3 gene and the proximal domino poly(A) site. These experiments showed a slight increase of RNA Pol II and Symplekin association with the histone gene and a modest decrease in association of these proteins with the domino 3’ end (Fig. 5B, left). CPSF73, one of the factors codepleted by CPSF160 knockdown showed a modest decrease in association with the 5’ end of histone H3 gene but a high level of association with the 3’ end of histone H3 gene (Fig. 5B, right, white bars). Strikingly, CPSF73 levels were down to 10% of maximum value in CPSF160 knockdown cells on the poly(A) site of the domino gene (Fig. 5B).
These data show that codepletion of Symplekin and CPSF73 by knockdown of polyadenylation factors (CstF64 and CPSF160, respectively) which are not required for histone 3’ end processing, does not affect recruitment of CPSF73 or Symplekin to histone genes, but it does affect recruitment to the domino gene. These data suggest that there are two distinct cleavage complexes each containing the same core proteins but requiring different accessory factors depending on the type of RNA to be processed. Knockdown of a factor not associated with the histone cleavage complex may codeplete the polyadenylation cleavage complex, but not the histone cleavage complex.
Discussion
All mRNAs in eukaryotic cells are transcribed by RNA Pol II and the first step in 3’ end formation of these mRNAs is endonucleolytic cleavage. Cleavage is coupled with polyadenylation for most genes, but cleavage is the only step in 3’ end formation of metazoan replication-dependent histone mRNAs. In the last few years, it has become apparent that there are factors shared between these two processes, and that CPSF73 catalyzes the endonucleolytic cleavage of both polyadenylated mRNAs (Mandel et al., 2006) and histone mRNAs (Dominski et al., 2005b). Here we provide evidence that a subset of poly(A) factors, CPSF73, CPSF100 and Symplekin, are present as a complex in Drosophila cells, and this complex likely forms the core of both the polyadenylation and histone cleavage factors.
CPSF73, CPSF100 and Symplekin comprise the mRNA processing core cleavage factor
Depletion of the cleavage and poly(A) factors CPSF73, CPSF100, and Symplekin from Drosophila cultured cells results in misprocessing of histone pre-mRNA. When SLBP or U7 snRNP components are RNAi depleted, most of the misprocessed histone mRNA is polyadenylated. This is in contrast to CPSF73, CPSF100 or Symplekin depletion where the majority of misprocessed mRNAs are run-on transcripts that are not polyadenylated (Fig. 1). These data are consistent with knockdown of these factors also causing polyadenylation defects in vivo (Wagner et al., 2007) and with a subset of poly(A) factors forming a complex required for both poly(A) and histone pre-mRNA processing.
We find that RNAi depletion of some factors results in codepletion of other proteins. Since RNAi does not significantly affect the mRNA levels of other components, we hypothesize that the codepleted factors are present in a complex that is destabilized by reduction of a single protein. For example, depletion of CPSF73, CPSF100 or Symplekin results in codepletion of the other two factors, but does not affect levels of SLBP, CstF50 or CstF77 (Fig. 2). CstF is a second complex that exhibits similar codepletion characteristics since CstF50, CstF64, and CstF77 are codepleted as a result of knockdown of any one of the three components. These data agree with the findings of Manley and coworkers (Hatton et al., 2000; Takagaki et al., 1990) in mammalian cells.
The presence of a stable complex containing CPSF73, CPSF100 and Symplekin is further supported by IP experiments using antibodies to CPSF73 and Symplekin under stringent buffer conditions. While substantial amounts of endogenous CPSF73, CPSF100 and Symplekin coprecipitate with one another (Fig. 2) significantly less SLBP or Lsm11 coimmunoprecipitated with Symplekin or CPSF73, although there was reproducible coimmunoprecipitation of small amounts of these endogenous factors under less stringent buffer conditions. This rare complex may represent a small fraction of cleavage factor that is transiently associated with histone processing factors. Previous studies of in vitro processing of histone pre-mRNA in Drosophila, demonstrated that both SLBP and U7 snRNP are essential for processing consistent with them both being required to recruit the cleavage factor (Dominski et al., 2005a)
The core cleavage factor is recruited to histone genes in vivo
Our current model of how the cleavage factors are recruited to histone genes is shown in figure 6. We determined that the core cleavage factor associates with nascent pre-mRNAs by performing ChIP assays using antibodies to RNA Pol II and components of the cleavage machinery. These results revealed that under normal conditions RNA Pol II does not proceed more than 500 nts past the 3’ end of the histone H3 mRNA, suggesting that processing occurs shortly after transcription of the HDE (Fig. 3). These results agree with the in vitro results of Adamson and Price (Adamson and Price, 2003) who detected strong pause sites just downstream of the HDE of all 5 Drosophila histone genes. Pausing of RNA Pol II may allow time for assembly of the processing complex and subsequent transcription termination. We also detected association of CPSF73 and Symplekin on the 3’ ends of histone genes (Figs. 3 and 6A), consistent with cotranscriptional formation of the processing complex and likely processing. CPSF73 has previously been found by ChIP to associate with the 3’ ends of mammalian histone genes (Glover-Cutter et al., 2008). Recent work suggests that CPSF73 acts as an exonuclease after processing to degrade the downstream cleavage product and possibly enhance termination on histone genes. This would require cotranscriptional processing of histone pre-mRNA (Yang et al., 2009).
Figure 6. Model of histone pre-mRNA processing.
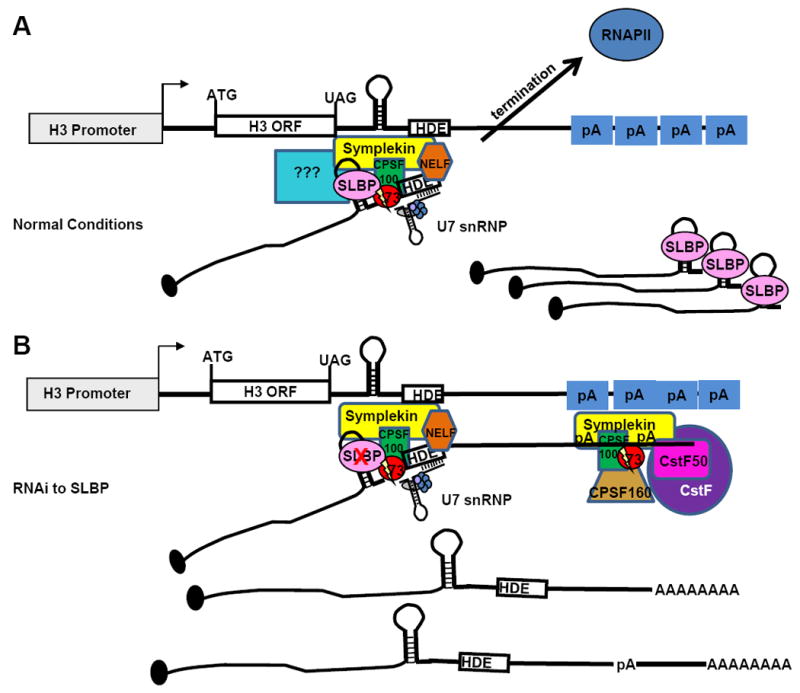
(A) Model of normal histone 3’ end formation. (B) Additional poly(A) factors are recruited to histone genes to participate in cleavage and polyadenylation in the absence of histone specific RNA processing proteins.
Notably, we did not detect large amounts of CstF50 on histone genes under normal conditions. However, when SLBP was RNAi depleted, RNA Pol II was detected 3’ of the normal processing site and CPSF73, Symplekin and CstF50 were now associated with histone genes (Fig. 4). These data are consistent with recruitment of additional poly(A) machinery to histone genes upon transcription to polyadenylation sites located downstream of the histone processing site (Fig. 6B).
Two distinct complexes, both containing the core cleavage factor, are required for histone mRNA and poly(A) mRNA 3’ end formation
Knockdown of CPSF160, a factor specific for polyadenylation, also causes depletion of Symplekin, CPSF-73 and CPSF-100 (Fig. 2), yet, this codepletion has no effect on histone pre-mRNA processing or on recruitment of Symplekin or CPSF-73 to the 3’ ends of histone genes (Fig. 5). As expected, knockdown of CPSF160 or CstF64 resulted in reduction of CPSF73 and Symplekin, respectively, associated with the 3’ of the domino gene (Fig. 5). These data support our hypothesis that the histone cleavage factor is distinct from the polyadenylation cleavage factor and that knockdown of CPSF160 or CstF64 codepletes the polyadenylation cleavage factor and not the histone cleavage factor. Thus the core cleavage factor containing CPSF73, CPSF100 and Symplekin likely associates with different accessory proteins to process either poly(A) or histone mRNAs (Fig. 2D).
A comparison of histone and poly(A) mRNA 3’ cis elements supports this model. Histone pre-mRNAs have two cis elements, the SL that binds SLBP and the HDE which base pairs to the U7 snRNA in the U7 snRNP. The SL and HDE are analogous to the AAUAAA and G/U rich downstream sequence element (DSE) in the 3’ UTRs of canonical poly(A) mRNAs. CPSF160 interacts with the AAUAAA and CstF64 binds the DSE. In both cases endonucleolytic cleavage is accomplished by the core cleavage factor consisting of CPSF73, CPSF100 and Symplekin.
It is likely that the cell maintains these two separate complexes and directs them to the proper targets. CPSF160 binding to the AAUAAA is sufficient for recruitment of CPSF73 and CPSF160 to the pre-mRNA although additional factors are required to activate cleavage. Concentration of the U7 snRNP in the HLB is critical for efficient histone mRNA processing in vivo (Wagner et al., 2007) and it is possible a local concentration of U7 snRNP in the HLB is critical for recognition of histone pre-mRNA and recruitment of the histone cleavage factor to histone genes.
Histone pre-mRNA 3’ end processing is cotranscriptional
The factors involved in transcription of histone genes include many proteins required for transcription of canonical polyadenylated mRNAs. Unlike mammalian snRNAs (Hernandez and Weiner, 1986), there is no evidence that histone mRNA 3’ formation end is affected by the promoter used for transcription as a histone reporter driven by the actin or globin promoter forms histone 3’ ends as effectively as one driven by the histone promoter. Similarly, a histone promoter driving a polyadenylated mRNA results in efficient polyadenylation [B. Burch, E.J. Wagner and W.F.M, unpublished and (Whitelaw et al., 1986)]. The ~400 nt coding regions of histone genes are highly conserved and factors which bind to the nascent transcript could help recruit processing factors (Friend et al., 2007).
Many factors involved in transcription and processing of canonical polyadenylated mRNAs via interaction with the CTD of RNA Pol II, such as NELF and the cap binding complex (Narita et al., 2007), are likely also recruited to histone genes. There is substantial evidence that some components of the poly(A) machinery can bind to the CTD of RNA Pol II [reviewed in (Buratowski, 2005; Neugebauer, 2002)] and coordinate transcription with 3’ end processing. For example, CstF interacts preferentially with the phosphorylated CTD (McCracken et al., 1997). The yeast orthologue of CPSF160 can also interact with the CTD directly (Dichtl et al., 2002). The interaction between the CTD and CstF may account for the small amount of occupancy of CstF50, a factor not involved in histone pre-mRNA processing, observed on histone genes (Fig. 3). Moreover, recent studies have suggested that there is a substantial difference in the modification of RNA Pol II in CBs (Xie and Pombo, 2006). We hypothesize that a high local concentration of histone pre-mRNA processing factors, possibly in concert with CTD recruitment of these factors, is critical for histone pre-mRNA processing. These data together with the studies of Adamson and Price, point to a major role for stalling of the RNA polymerase II and binding of the factors to the nascent RNA. It seems likely that the histone cleavage factor is not directly associated with the CTD but more likely is recruited as a result of SLBP and U7 snRNP to the nascent transcript.
MATERIALS AND METHODS
RNAi and NE preparation
~500 bp dsRNAs were made by T7 transcription. RNAi was performed by adding 100 μg of dsRNA to Dmel-2 cells (2 × 107) on day 1, 200 μg on day 2 and 400 μg on day 3. Following a two-day recovery, NEs were prepared as described (Wagner et al., 2007).
Antibodies
Amino acids 1-767 of D. melanogaster Symplekin and full length CPSF73 were expressed in E.coli and used to inoculate rabbits and guinea pigs for antibody production (Pacific Immunology Corp.). Amino acids 1-175 of D. melanogaster SLBP or amino acids 1-100 of Lsm11 were used to inoculate rabbits for antibody production (Proteintech Group, Inc.). The D. melanogaster α-CstF50 antibody (Ni et al., 2008) was a gift from John Lis (Cornell Univ.) and the α–Rpb3 antibody (Gilchrist et al., 2008) was a gift from Karen Adelman (NIEHS). The α-CstF77 antibody was from Bethyl Labs. The CPSF-100 antibody was raised against a conserved region of human CPSF-100 (Z. Dominski and X.Yang, unpublished) and cross-reacted with the D. melanogaster CPSF-100 protein.
Analysis of RNA
The S1 nuclease assay was performed as described (Lanzotti et al., 2002) . Poly(A) mRNA was separated from total cellular RNA using the Poly(A)Purist mRNA isolation kit (Ambion).
Immunoprecipitations
500 μg NE were incubated with pre-conjugated protein-A sepharose beads in RIPA (50 mM Tris, pH 8.0, 1% NP-40, 0.25% deoxycholate, 150 mM NaCl) or a low detergent buffer (50 mM Tris, pH 8.0, 0.1% NP40, 150 mM NaCl). Beads were washed in IP buffer and the precipitated proteins were eluted into SDS loading buffer.
Chromatin Immunoprecipitation
ChIP was performed as described (Gilchrist et al., 2008) and 15 μl of CPSF73, Rpb3, Symplekin and CstF50 antibodies were used in each IP. qPCR was performed and quantified using SYBR green (Applied Biosystems) and an Applied Biosystems 7900HT real-time PCR machine.
Immunofluorescence
IF was performed as described (Wagner et al., 2007) using α-Mpm-2 (Sigma) and α-CstF50 primary antibodies.
Supplementary Material
Acknowledgments
This work supported by NIH grant GM58921 to W.F.M. K.D.S was supported by an NIH training grant (T32GM008581) and M.S. was supported by an NIH training grant (T32CA09156) and NIH fellowship (F32GM080950). We thank Karen Adelman (NIEHS, RTP, NC) for advice on the ChIP assays and the RPB3 antibody and John Lis (Cornell Univ. Ithaca, NY) for the CstF50 antibody.
Footnotes
Supplemental Data
Supplemental data includes materials and methods and one figure.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson TE, Price DH. Cotranscriptional processing of Drosophila histone mRNAs. Mol Cell Biol. 2003;23:4046–4055. doi: 10.1128/MCB.23.12.4046-4055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3’ end processing and transcription termination. Curr Opin Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang X, Purdy M, Marzluff WF. Differences and similarities between Drosophila and mammalian 3’ end processing of histone pre-mRNAs. RNA. 2005a;11:1835–1847. doi: 10.1261/rna.2179305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone pre-mRNA processing. Cell. 2005b;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7 snRNP dependent 3’ end formation. Molecular Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O, Krämer A, Vasserot A, Birnstiel ML. Heat-labile regulatory factor is required for 3’ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and block oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton LS, Eloranta JJ, Figueiredo LM, Takagaki Y, Manley JL, O’Hare K. The Drosophila homologue of the 64 kDa subunit of cleavage stimulation factor interacts with the 77 kDa subunit encoded by the suppressor of forked gene. Nucleic Acids Res. 2000;28:520–526. doi: 10.1093/nar/28.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM. Formation of the 3’ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3’ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon BH, Schafer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3’-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3’-end maturation. EMBO Rep. 2008 doi: 10.1038/embor.2008.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes and Development. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Sadowski M, Dichtl B, Keller W. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3’-end formation. Nucleic Acids Res. 2003;31:3936–3945. doi: 10.1093/nar/gkg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3’ processing in vivo. Mol Cell Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3’ end processing factors with RNA polymerase II. Molecular Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3’-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan GH, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Murthy KGK, Manley JL. The 160-kD subunit of human cleavage polyadenylation specificity factor coordinates pre-mRNA 3’-end formation. Genes and Development. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF Interacts with CBC and Participates in 3’ End Processing of Replication-Dependent Histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor JP, Peebles CL. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Identifying PhosphoCTD-Associating Proteins. Methods Mol Biol. 2004;257:17–28. doi: 10.1385/1-59259-750-5:017. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3’ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3’ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA. 2009;15:459–472. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3’ processing complex. Mol Cell Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzer HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Molecular Cell. 2007;28:692–699. doi: 10.1016/j.molcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Coates A, Proudfoot NJ. Globin gene transcripts can utilize histone gene 3’ end processing signals. Nucleic Acids Res. 1986;14:7059–7070. doi: 10.1093/nar/14.17.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SQ, Pombo A. Distribution of different phosphorylated forms of RNA polymerase II in relation to Cajal and PML bodies in human cells: an ultrastructural study. Histochem Cell Biol. 2006;125:21–31. doi: 10.1007/s00418-005-0064-2. [DOI] [PubMed] [Google Scholar]
- Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies on the 5’ exonuclease and endonuclease activity of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol. 2009;29:21–32. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshio TD, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kessler M, Helmling S, O’Connor JP, Moore C. Pta1, a component of yeast CFII, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


