Figure 4. Loss of a histone specific processing factor results in recruitment of additional poly(A) factors to histone genes.
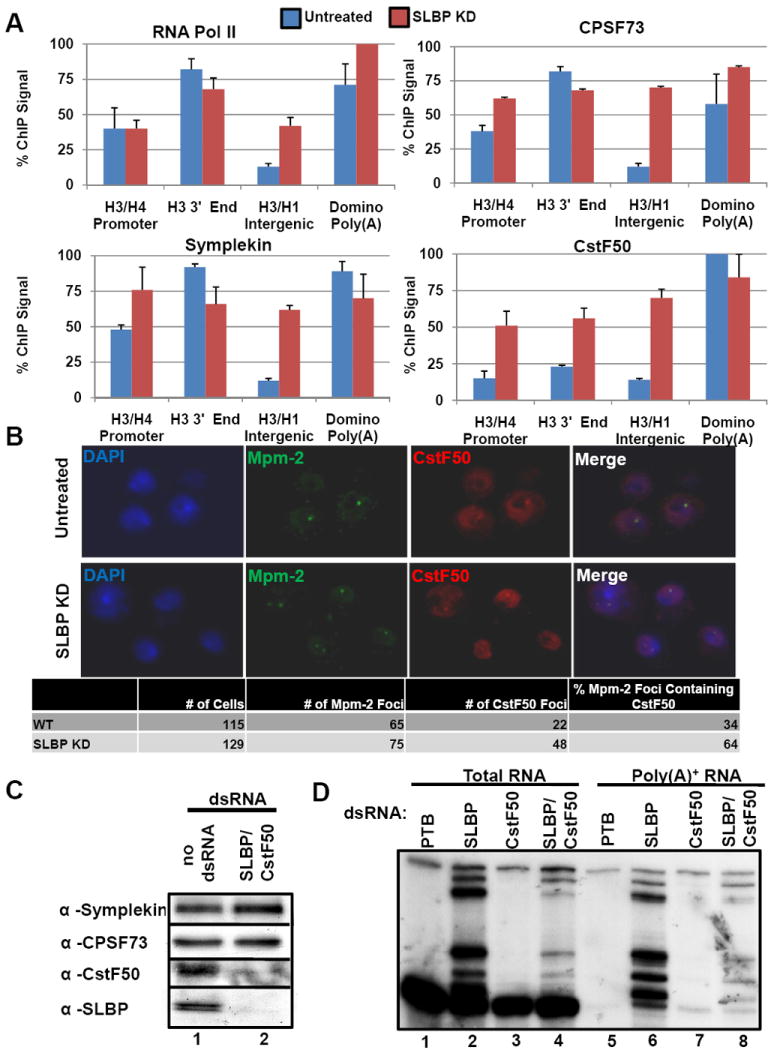
(A) ChIP assays were performed using lysates prepared from Dmel-2 cells treated with control (blue) or SLBP (red) dsRNAs. Columns represent the average relative ChIP signal normalized to the maximal value for each antibody. Data are presented as the average of 3 independent experiments +SD. Blue bars are untreated cells; red bars are values for SLBP knockdown cells. (B) Dmel-2 cells treated with control dsRNAs (top) or SLBP dsRNAs (bottom) were fixed and probed for CstF50 (green) and Mpm-2 (red), and also stained with DAPI (blue). The number of cells with Mpm-2 foci, and the number of cells with Mpm-2 foci that also contained CstF50 foci were counted. (C) Western blots of Dmel-2 cells treated with PTB dsRNA (control) or dsRNA targeting SLBP and CstF50. (D) Total RNA was prepared from cells treated as in (C) or with dsRNAs against SLBP or CstF50. S1 nuclease protection assays were performed on total RNA (lanes 1-4) or poly(A)+ RNA (lanes 5-8) as in Fig. 1 C, D.
