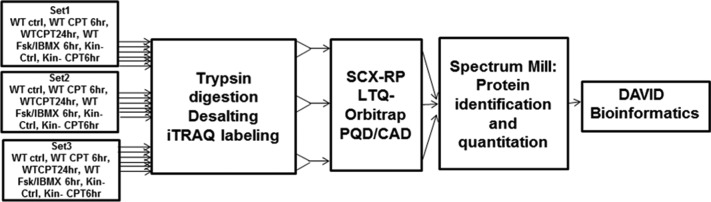
Figure 1.
Diagram of the workflow of the proteomic studies. Proteins in wild-type (WT) and kin- S49 cells were digested in three biological replicates of cells (WT control, WT for 6 and 24 h with CPT-cAMP (100 μM) treatment, WT incubated with 100 μM IBMX and 10 μM Fsk, kin- control, and kin- cells incubated with 100 μM CPT-cAMP for 6 h). The resulting peptides were desalted and labeled with different iTRAQ reagents. The labeled peptides from each sample were pooled and then separated with online 2D LC with PQD/CAD acquisition on an LTQ-Orbitrap. Spectrum Mill was used to search the raw data for protein identification and quantitation. The proteins whose level of expression was increased or decreased were analyzed using the DAVID bioinformatics tool.