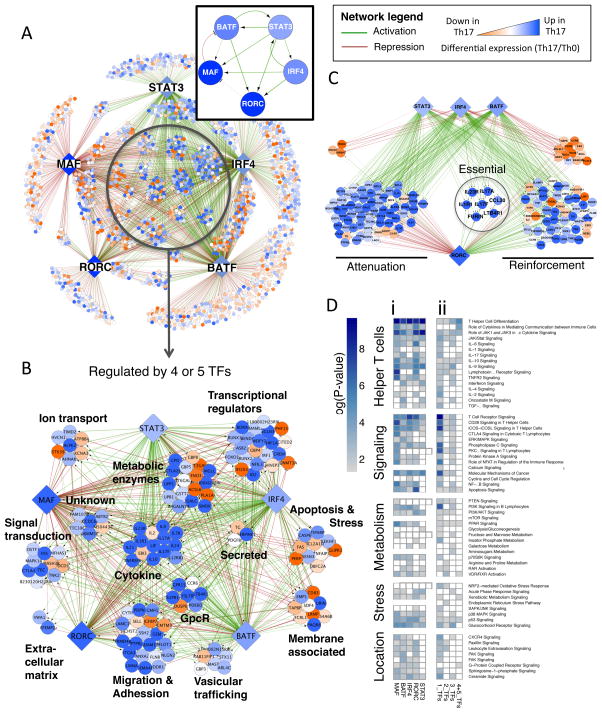
Figure 3. TF combinatorial interactions specify the Th17 lineage.
A. High confidence regulatory edges (FDR<10%; based on 10,000 simulations) focused on five core TFs identify direct (ChIP-seq) and functional (KO RNA-seq) regulatory targets (visualized using Cytoscape). Boxed inset displays the regulatory interactions between core Th17 TFs. See network legend for visualization scheme.
B. Expanded view of highly regulated nodes with four to five core regulatory inputs, grouped based on general functional categories.
C. Regulatory interactions shared by STAT3, IRF4, BATF, and RORγt highlighting different aspects of RORγt transcriptional function. Attenuation: RORγt repression targets that are up-regulated in Th17 cells; Reinforcement: Activation targets that are up-regulated in Th17 cells; Essential: targets having a two-fold change in RORγt KO differential expression and KO H3K4me3 ChIP.
D. Targets for single TFs are enriched for pathways in multiple functional categories (i). Targets of multiple TFs (increasingly regulated by 2, 3, or 4+5 TFs) are selectively enriched for pathways related to T helper differentiation and effector function (ii). Analysis performed using the Ingenuity analysis tool (IPA) is presented as a heat map of enrichment p-values.