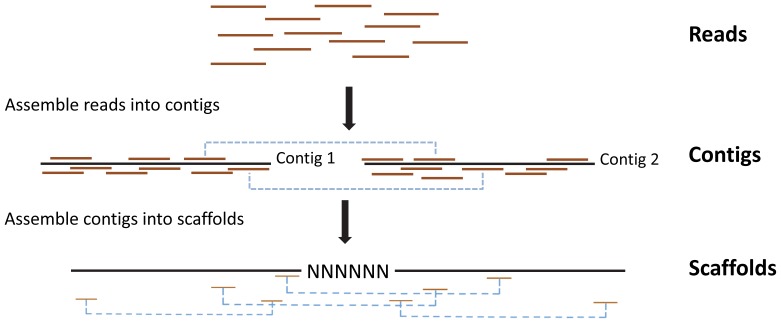
Figure 2. Schematic representation of the method used to assemble Illumina reads into contigs, and contigs into scaffolds.
All reads were initially assembled into contigs using the de Bruijn graph method without using information about paired-end reads (shown by blue dashed lines). A contig’s sequence was resolved at every base. Contigs were then assembled into longer scaffolds by connecting contigs that contained paired-end reads assembled into separate contigs. Assembling scaffolds in this way allowed us to create longer sequences of known length, but sometimes there were gaps of unknown sequence. These gaps were constrained to represent <5% of total sequence length.