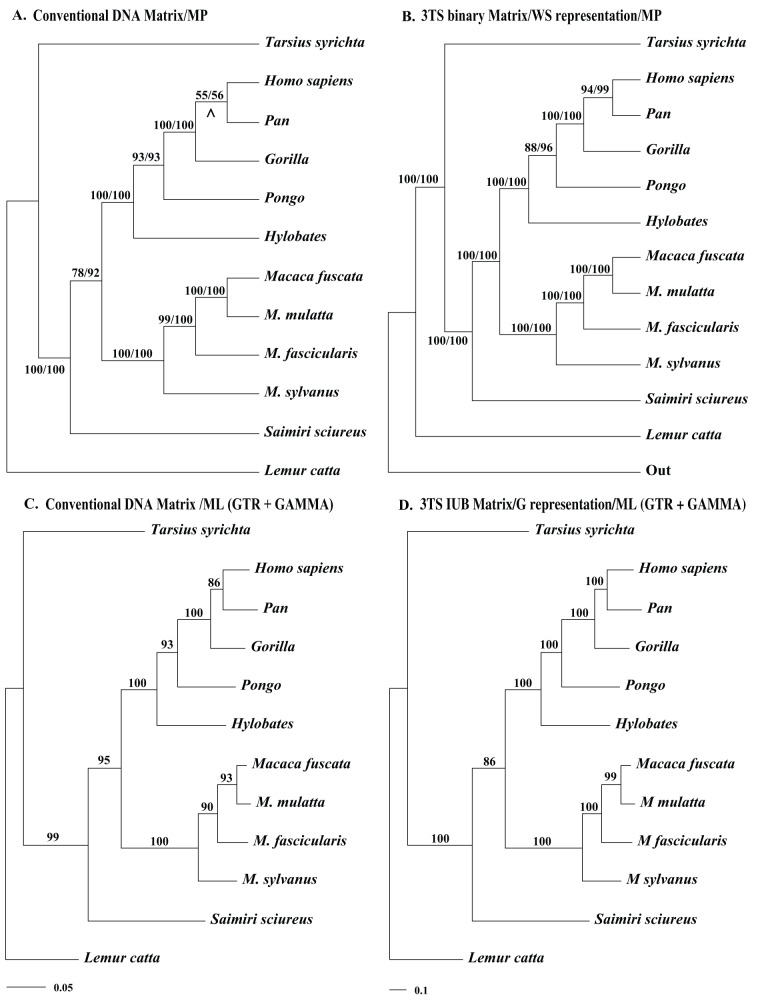
Figure 1. Example Maximum Parsimony.
(MP) and Maximum Likelihood (ML) analyses of the conventional and 3TS represented clock-like umc-matrices. A. One of the two most parsimonious trees (length = 1153, CI = 0.6496, RI = 0.5960) recovered from a MP analysis of the clocklike mt NADH-dehydrogenase subunit-4 (NADH-4) (Hayaska et al., 1988, summarized in [15]). The matrix comprised a total of 898 nucleotide characters, of which 367 were parsimony informative. All characters were treated as “unordered” (Fitch parsimony). The MP analysis was conducted in PAUP* 4.0b10 [15], with an heuristic search with 1000 random addition replicates (saving no more than 100 trees per replicate), and the TBR branch swapping/MulTrees option in effect [15]; gaps were treated as “missing” data. Branches with a minimum length of zero were collapsed. Bootstrap (MP BS)/Jackknife (MP JK) values, respectively, are provided above or below the branches, and indicate nodes with greater than 50% support. Both MP BS/JK estimates were obtained using 100 replicates and 100 random addition sequences (saving no more than 1000 trees per replicate), with the TBR branch swapping/MulTrees option in effect. In total, 36.0% of the characters were deleted in each JK replicate. Lemour catta selected as the outgroup. B. The single, most parsimonious tree (length = 26305, CI = 0.8142, RI = 0.07718). recovered from an MP analysis of a 3TS binary matrix generated by TAXODIUM from the NADH-4 matrix (Figure 1A). See the Figure 1A legend for the details of MP analysis. Williams-Siebert (WS) representation was performed, and the value of the outgroup was fixed as Lemour catta; all 3TS were uniformly weighted (command: taxodium.exe input_file_name.csv –idna –ob –og –nex). The number of characters (3TSs) was 21418, of which all were parsimony informative. C. The Maximum Likelihood (ML) tree (log likelihood = −5722.341070) inferred from the NADH-4 matrix (Figure 1A) with RAxML [16]. A total of 500 rapid bootstrap (ML BS) replicates were used to assess support for individual nodes. GTR+ G substitution model assumed as a best choice. D. The ML tree (log likelihood = −208795.522546) inferred from the IUB-notated 3TS matrix generated by TAXODIUM from the NADH-4 matrix ( Figures 1A, C). See the Figure 1C legend for additional details of ML analysis. G-representation was performed; an operational outgroup was not explicitly defined and, therefore, was not included in the analysis (command: taxodium.exe input_file_name.csv –idna –odna –phy). The total number of characters (3TSs) in the 3TS matrix was 61043.