Table 1.
Rate and equilibrium constants for glmS ribozyme reactions with different cofactors.
| Cofactor | Structure |
kobs (min−1) 10 mM, pH 7.5, 25 °C |
Kd,app (mM) pH 7.5, 25 °C |
kcat/Km (mM−1min−1) pH 7.5, 25 °C |
|---|---|---|---|---|
| D-glucosamine-6-phosphate (GlcN6P) |
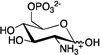 |
92.9 ± 5.3 | 0.96 ± 0.11 | 98 ± 5 |
| D-glucosamine-6-sulfate (GlcN6S) |
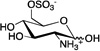 |
3.1 ± 0.6 | 270 ± 64 | 0.22 ± 0.01 |
| D-glucosamine (GlcN) |
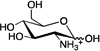 |
0.65 ± 0.01 | ~ 500 | 0.062 ± 0.001 |
| serinol (2-amino-propane-1,3-diol) |
0.013 ± 0.002 | ≥ 1000 | 0.00047 ± 0.00007 | |
| TRIS (2-amino-2-hydroxymethyl- propane-1,3-diol) |
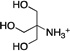 |
0.0096 ± 0.0006 | ≥ 1000 | 0.00035 ± 0.00014 |
| L-serine | 0.0095 ± 0.0001 | ≥ 1000 | 0.00029 ± 0.00005 | |
| ethanolamine | 0.003 ± 0.001 | ≥ 1000 | 0.000058 ± 0.000003 | |
| *L-prolinol | 0.0007 ± 0.0003 | ≥ 1000 | 0.000023 ± 0.000009 | |
| no cofactor | - | 0.0001 | -- | -- |
not previously described
