Figure 5.
Loss of Piwi, but Not Mael, Leads to Decreased H3K9me3 on TEs
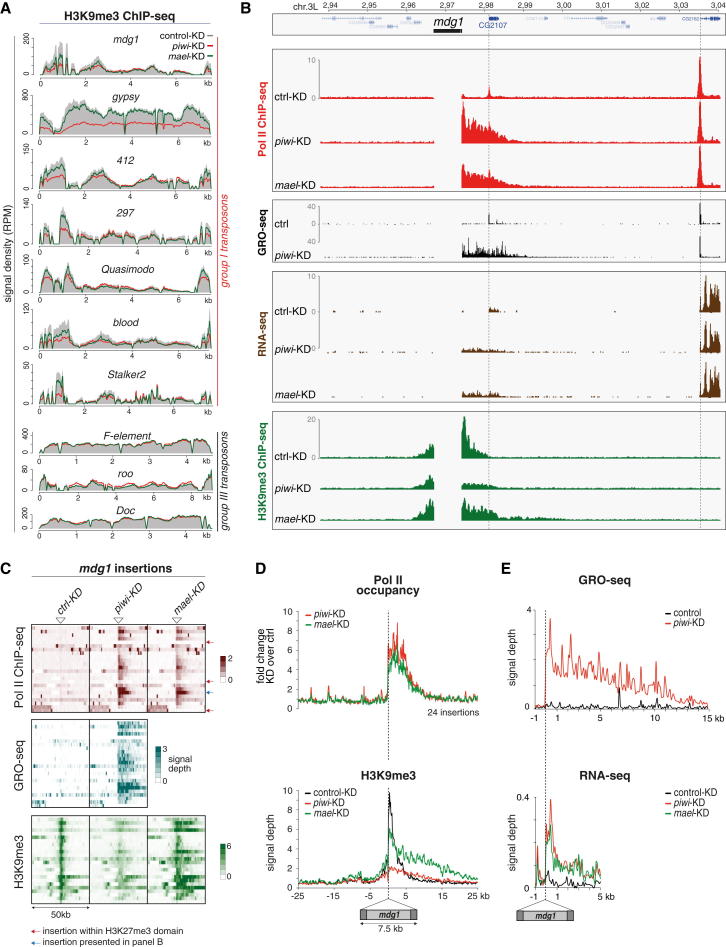
(A) Profiles of normalized H3K9me3 signals on indicated TE consensus sequences (group I versus group III) in piwi KD cells (red line), mael KD cells (green line), and GFP KD cells (solid gray).
(B) Profiles of transcriptional activity (Pol II ChIP-seq, GRO-seq, RNA-seq) and H3K9me3 density of OSCs after indicated knockdowns (left) in a 100 kb window (chr. 3L; 63E) that contains an mdg1 insertion (only genome unique reads are displayed).
(C) Heatmaps presenting Pol II occupancy, GRO-seq signal, and H3K9me3 ChIP-seq signal within 50 kb windows centered on all 24 euchromatic mdg1 insertions (oriented 5′ to 3′) in control, piwi, or mael KD cells. Red arrows mark insertions within H3K27me3 domains; the insertion marked by the blue arrow is the one displayed in (B).
(D and E) Metaplots for the genomic regions flanking the average euchromatic mdg1 insertion; displayed are changes in Pol II occupancy (D), normalized H3K9me3 signal (D), normalized GRO-seq signal (E), and normalized RNA-seq signal (E) in control KD (black), piwi KD (red), or mael KD cells (green).
See also Figure S4.