Figure 6.
Euchromatic H3K9me3 Islands Correlate with TE Insertions and Depend on Piwi
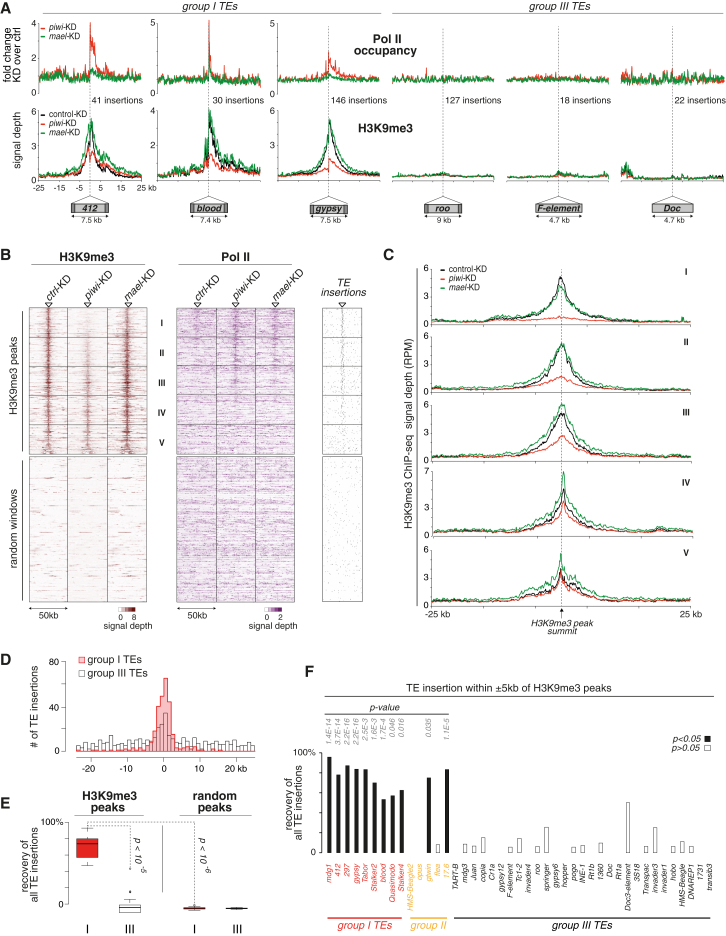
(A) Metaplots for the genomic regions flanking the average euchromatic insertion of indicated TEs (copy number indicated) belonging to group I or III; displayed are changes in Pol II occupancy (compared to control KD) and normalized H3K9me3 signal in control KD (black), piwi KD (red), or mael KD cells (green).
(B) Heatmaps showing signal of H3K9me3 (left) or Pol II occupancy (right) in 50 kb windows centered on euchromatic H3K9me3 peaks (n = 466) and in a random set of euchromatic 50 kb windows (n = 466) in control KD, piwi KD, or mael KD cells. Windows with euchromatic H3K9me3 peaks were sorted according to the loss of signal in piwi KD cells, and five equally sized bins were defined (I–V). To the right, positions of all TE insertions within the euchromatic H3K9me3 windows are displayed.
(C) Metaplots of H3K9me3 signals (individual plots for the five bins defined in B) for the 50 kb window flanking the average euchromatic H3K9me3 peak in control (black), piwi KD (red), or mael KD cells (green).
(D) Distribution of group I (red) or III (white) TE insertions in the 50 kb windows centered on all H3K9me3 peaks (n = 466).
(E) Percentages of all group I (red) or all group III (white) TE insertions found within 5 kb of euchromatic H3K9me3 summits in comparison to an average and randomly selected set of control regions. Box plots are as in Figure 3.
(F) Percentages of insertions of indicated individual TEs found within 5 kb of euchromatic H3K9me3 summits (p values based on binominal test using random control areas).