Figure 7.
The Impact of TEs and the piRNA Pathway on Gene Expression
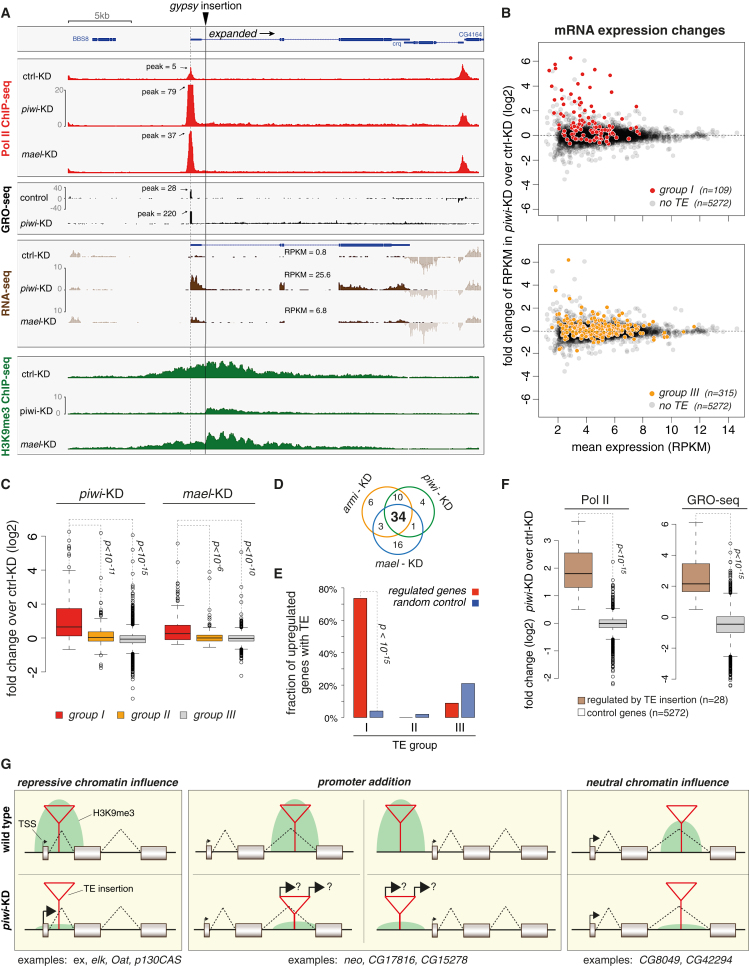
(A) Transcriptional activity (Pol II ChIP-seq, GRO-seq, RNA-seq) and H3K9me3 density at the expanded locus. OSC-specific insertion site of gypsy indicated with gray line; ex TSS, with dashed line.
(B) Changes in mRNA abundance for the set of expressed genes (RPKM >5 in one library) between piwi KD and control KD cells. The upper plot contrasts genes with no insertion (gray) and genes with an insertion of a group I TE in sense orientation (red). The lower plot contrasts genes with no insertion (gray) and genes with no group I TE but with a group III TE insertion (yellow).
(C) Box plot showing the changes (log2) in RNA-seq RKPM levels for the three gene groups defined in (B) upon knockdown of Piwi (left) or Mael (right).
(D) Venn diagram displaying the number of upregulated genes upon depletion of the indicated piRNA pathway components (criteria: armi and piwi KDs >4-fold, mael KD >2-fold; compared to GFP KD).
(E) Shown is the percentage of all upregulated genes (red) with TE insertions belonging to class I, II, or III in close proximity (± 5 kb) in comparison to random control genes (blue).
(F) Box plots displaying fold changes (log2) in Pol II occupancy (left) or GRO-seq signal (right) upon piwi KD versus GFP KD for upregulated genes with TE insertion (n = 28) versus all expressed genes with no TE insertion (n = 5272; RPKM >5). Box plots are as in Figure 3.
(G) Cartoon showing the three categories of how TE insertions impact gene loci. For category A (repressive chromatin influence), the TE insertion is close to the gene's TSS and results in TSS repression via H3K9me3 spreading. For category B (promoter addition), the TE insertion serves as an ectopic promoter through transcriptional bleeding. For category C (neutral chromatin influence), the TE insertion does not dampen gene expression but triggers local H3K9me3 spreading. Examples for each category are given below.
See also Figure S5.