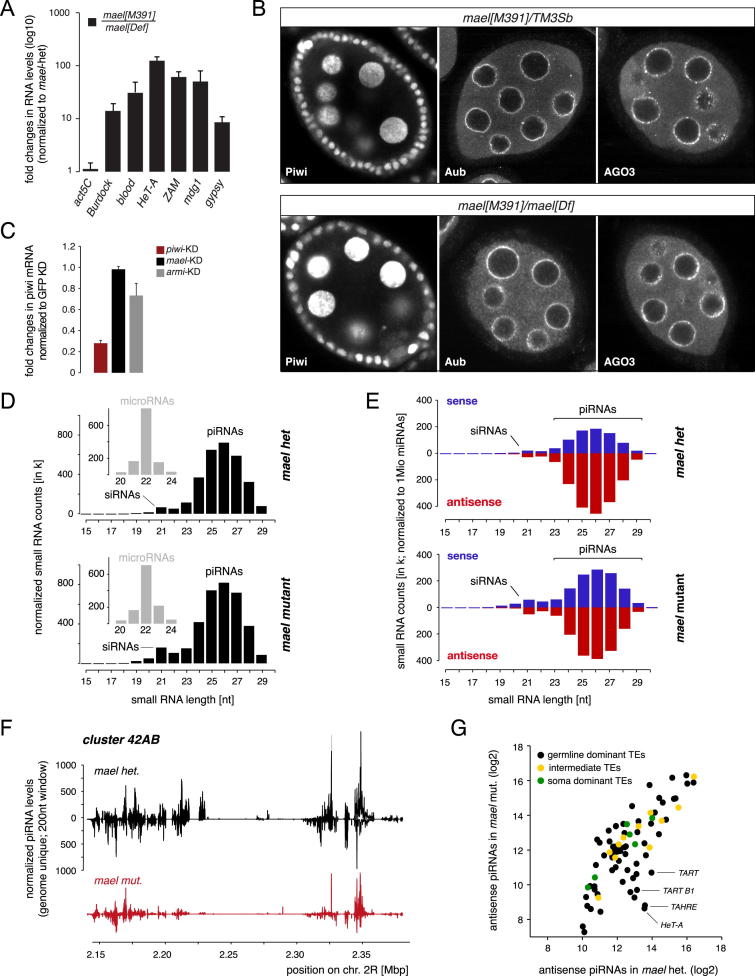
Figure S1.
Related to Figure 1
(A) Displayed are fold changes in steady state RNA levels of indicated TEs in mael null mutant ovaries (normalized to mael heterozygote siblings; values are averages of 3 biological replicates (error bars: StDev.).
(B) Confocal images of mael heterozygous (top) or mael[M391]/mael[Def] (bottom) egg chambers stained for Piwi, Aub or AGO3.
(C) Displayed are fold changes in piwi steady state mRNA levels in OSCs after transfection with indicated siRNAs. Values are averages of 3 biological replicates (error bars: StDev.) and normalized to GFP siRNA treated cells.
(D) Shown are length profiles of small RNAs (normalized to 1 million microRNAs; small insets) isolated from ovaries of mael[M391] heterozygous or mael[391]/mael[Def] flies. siRNA and piRNA populations are indicated.
(E) Shown are length profiles of repeat derived ovarian small RNAs (normalized to 1Mio miRNAs) from mael heterozygous and mael[M391]/mael[Def] flies. (red antisense; blue sense). (F) Normalized piRNA profiles (sense up; antisense down; 200nt windows) from mael het. (black) or mael mut. (red) libraries mapping uniquely to the 42AB piRNA cluster.
(G) Scatter plot (log2 scale) showing levels of antisense piRNAs mapping to soma dominant (green), intermediate (yellow) or germline dominant (black) TEs in mael het. or mael mut. libraries.