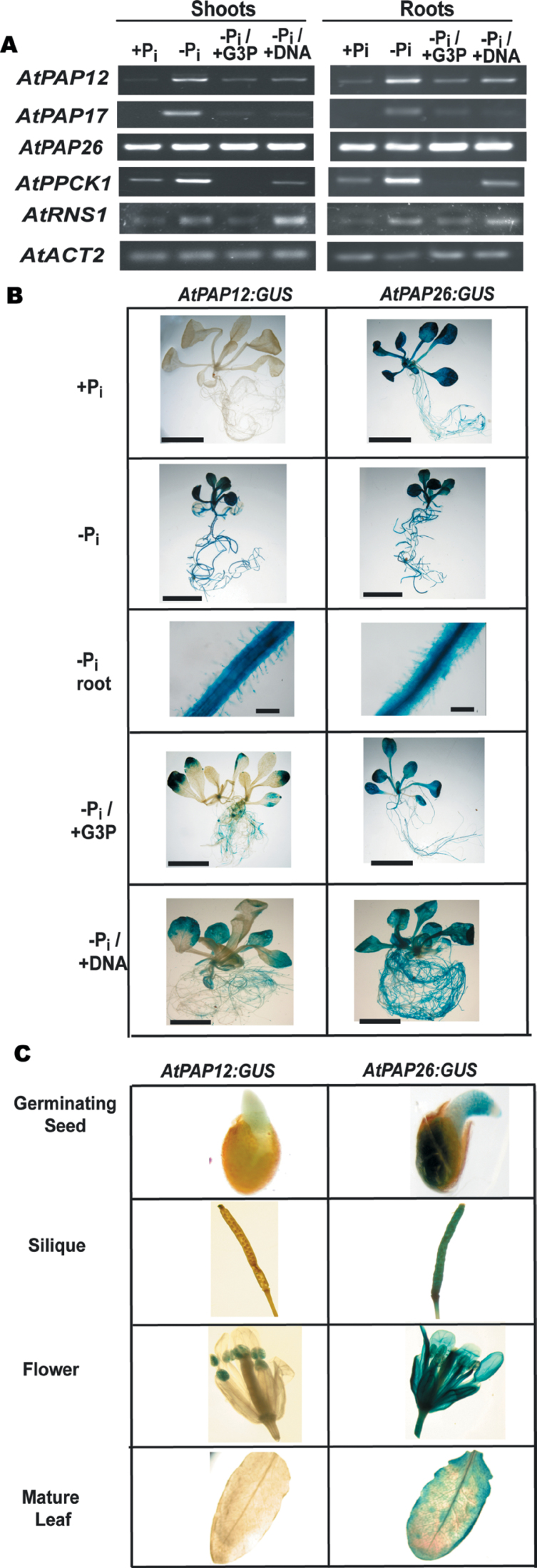
Fig. 2.
Analysis of AtPAP12 and AtPAP26 gene expression. (A) Levels of mRNA were analysed by semi-quantitative RT-PCR using gene-specific primers for AtPAP12, AtPAP17, AtPAP26, RNS1, and AtPPCK1. AtACT2 was used as a reference to ensure equal template loading. Seedlings were cultivated as described in the legend for Fig. 1. All PCR products were taken at cycle numbers determined to be non-saturating. Control RT–PCRs lacking reverse transcriptase did not produce any bands. (B) AtPAP12:GUS and AtPAP26:GUS transgenic lines were cultivated in 24-well microtitre plates in liquid MS medium containing 0.2mM Pi for 7 d, before being transferred into medium containing 0 or 1.5mM KH2PO4 (–Pi and +Pi, respectively), 1.5mM G3P (–Pi/+G3P), or 0.6mg ml–1 DNA (–Pi/+DNA) for another 7 d. Bars, 1cm, except for ‘–Pi root’ (bar, 100 µm). (C) AtPAP12:GUS and AtPAP26:GUS expression was also examined in several aerial tissues of 4-week-old +Pi plants that had been cultivated in soil under a regular light/dark diurnal cycle. ‘Germinating seed’ is a representative image of seeds that had been placed on moist filter paper and allowed to germinate for 1 d before GUS staining.